The dominant Australian community-acquired methicillin-resistant Staphylococcus aureus clone ST93-IV [2B] is highly virulent and genetically distinct
- PMID: 21991381
- PMCID: PMC3185049
- DOI: 10.1371/journal.pone.0025887
The dominant Australian community-acquired methicillin-resistant Staphylococcus aureus clone ST93-IV [2B] is highly virulent and genetically distinct
Abstract
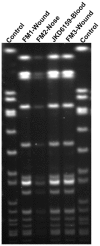
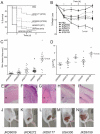
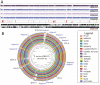
Community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) USA300 has spread rapidly across North America, and CA-MRSA is also increasing in Australia. However, the dominant Australian CA-MRSA strain, ST93-IV [2B] appears distantly related to USA300 despite strikingly similar clinical and epidemiological profiles. Here, we compared the virulence of a recent Australian ST93 isolate (JKD6159) to other MRSA, including USA300, and found that JKD6159 was the most virulent in a mouse skin infection model. We fully sequenced the genome of JKD6159 and confirmed that JKD6159 is a distinct clone with 7616 single nucleotide polymorphisms (SNPs) distinguishing this strain from all other S. aureus genomes. Despite its high virulence there were surprisingly few virulence determinants. However, genes encoding α-hemolysin, Panton-Valentine leukocidin (PVL) and α-type phenol soluble modulins were present. Genome comparisons revealed 32 additional CDS in JKD6159 but none appeared to encode new virulence factors, suggesting that this clone's enhanced pathogenicity could lie within subtler genome changes, such as SNPs within regulatory genes. To investigate the role of accessory genome elements in CA-MRSA epidemiology, we next sequenced three additional Australian non-ST93 CA-MRSA strains and compared them with JKD6159, 19 completed S. aureus genomes and 59 additional S. aureus genomes for which unassembled genome sequence data was publicly available (82 genomes in total). These comparisons showed that despite its distinctive genotype, JKD6159 and other CA-MRSA clones (including USA300) share a conserved repertoire of three notable accessory elements (SSCmecIV, PVL prophage, and pMW2). This study demonstrates that the genetically distinct ST93 CA-MRSA from Australia is highly virulent. Our comparisons of geographically and genetically diverse CA-MRSA genomes suggest that apparent convergent evolution in CA-MRSA may be better explained by the rapid dissemination of a highly conserved accessory genome from a common source.
Conflict of interest statement
Figures
References
-
- Moran GJ, Krishnadasan A, Gorwitz RJ, Fosheim GE, McDougal LK, et al. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355:666–674. - PubMed
-
- Voyich JM, Otto M, Mathema B, Braughton KR, Whitney AR, et al. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J Infect Dis. 2006;194:1761–1770. - PubMed
-
- Labandeira-Rey M, Couzon F, Boisset S, Brown EL, Bes M, et al. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science. 2007;315:1130–1133. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

