Human cancer long non-coding RNA transcriptomes
- PMID: 21991387
- PMCID: PMC3185064
- DOI: 10.1371/journal.pone.0025915
Human cancer long non-coding RNA transcriptomes
Abstract
Once thought to be a part of the 'dark matter' of the genome, long non-coding RNAs (lncRNAs) are emerging as an integral functional component of the mammalian transcriptome. LncRNAs are a novel class of mRNA-like transcripts which, despite no known protein-coding potential, demonstrate a wide range of structural and functional roles in cellular biology. However, the magnitude of the contribution of lncRNA expression to normal human tissues and cancers has not been investigated in a comprehensive manner. In this study, we compiled 272 human serial analysis of gene expression (SAGE) libraries to delineate lncRNA transcription patterns across a broad spectrum of normal human tissues and cancers. Using a novel lncRNA discovery pipeline we parsed over 24 million SAGE tags and report lncRNA expression profiles across a panel of 26 different normal human tissues and 19 human cancers. Our findings show extensive, tissue-specific lncRNA expression in normal tissues and highly aberrant lncRNA expression in human cancers. Here, we present a first generation atlas for lncRNA profiling in cancer.
Conflict of interest statement
Figures
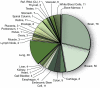
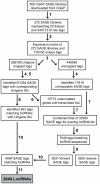

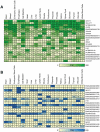
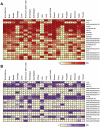
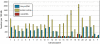
References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

