Genetic evolution of low pathogenecity H9N2 avian influenza viruses in Tunisia: acquisition of new mutations
- PMID: 21992186
- PMCID: PMC3223530
- DOI: 10.1186/1743-422X-8-467
Genetic evolution of low pathogenecity H9N2 avian influenza viruses in Tunisia: acquisition of new mutations
Abstract
Background: Since the end of 2009, H9N2 has emerged in Tunisia causing several epidemics in poultry industry resulting in major economic losses. To monitor variations of Influenza viruses during the outbreaks, Tunisian H9N2 virus isolates were identified and genetically characterized.
Methods: The genomic RNA segments of Tunisian H9N2 strains were subjected to RT-PCR amplifications followed by sequencing analysis.
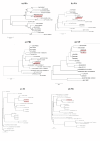
Results: Phylogenetic analysis demonstrated that A/Ck/TUN/12/10 and A/Migratory Bird/TUN/51/10 viruses represent multiple reassortant lineages, with genes coming from Middle East strains, and share the common ancestor Qa/HK/G1/97 isolate which has contributed internal genes of H5N1 virus circulating in Asia. Some of the internal genes seemed to have undergone broad reassortments with other influenza subtypes. Deduced amino acid sequences of the hemagglutinin (HA) gene showed the presence of additional glycosylation site and Leu at position 234 indicating to binding preference to α (2, 6) sialic acid receptors, indicating their potential to directly infect humans. The Hemagglutinin cleavage site motif sequence is 333 PARSSR*GLF341 which indicates the low pathogenicity nature of the Tunisian H9N2 strains and the potential to acquire the basic amino acids required for the highly pathogenic strains. Their neuraminidase protein (NA) carried substitutions in the hemadsorption (HB) site, similar to those of other avian H9N2 viruses from Asia, Middle Eastern and human pandemic H2N2 and H3N2 that bind to α -2, 6 -linked receptors. Two avian virus-like aa at positions 661 (A) and 702 (K), similar to H5N1 strains, were identified in the polymerase (PB2) protein. Likewise, matrix (M) protein carried some substitutions which are linked with increasing replication in mammals. In addition, H9N2 strain recently circulating carried new polymorphism, "GSEV" PDZ ligand (PL) C-terminal motif in its non structural (NS) protein.Two new aa substitutions (I) and (V), that haven't been previously reported, were identified in the polymerase and matrix proteins, respectively. Nucleoprotein and non-structural protein carried some substitutions similar to H5N1 strains.
Conclusion: Considering these new mutations, the molecular basis of tropism, host responses and enhanced virulence will be defined and studied. Otherwise, Continuous monitoring of viral genetic changes throughout the year is warranted to monitor variations of Influenza viruses in the field.
Figures
References
-
- Lamb RA, Krug RM. In: Fields Vrology. Knipe DM, Howley PM, Chanock RM, Melnick JL, Momath TP, Roizman S, editor. Philadelphia Lippincott Williams & Wilkins; 2001. Orthomyxoviridae: the viruses and their replication; pp. 1487–1532.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

