Self-replication of information-bearing nanoscale patterns
- PMID: 21993758
- PMCID: PMC3192504
- DOI: 10.1038/nature10500
Self-replication of information-bearing nanoscale patterns
Abstract
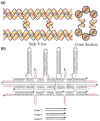
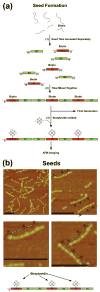
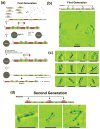
DNA molecules provide what is probably the most iconic example of self-replication--the ability of a system to replicate, or make copies of, itself. In living cells the process is mediated by enzymes and occurs autonomously, with the number of replicas increasing exponentially over time without the need for external manipulation. Self-replication has also been implemented with synthetic systems, including RNA enzymes designed to undergo self-sustained exponential amplification. An exciting next step would be to use self-replication in materials fabrication, which requires robust and general systems capable of copying and amplifying functional materials or structures. Here we report a first development in this direction, using DNA tile motifs that can recognize and bind complementary tiles in a pre-programmed fashion. We first design tile motifs so they form a seven-tile seed sequence; then use the seeds to instruct the formation of a first generation of complementary seven-tile daughter sequences; and finally use the daughters to instruct the formation of seven-tile granddaughter sequences that are identical to the initial seed sequences. Considering that DNA is a functional material that can organize itself and other molecules into useful structures, our findings raise the tantalizing prospect that we may one day be able to realize self-replicating materials with various patterns or useful functions.
Figures
References
-
- Wintner EA, Conn MM, Rebek J., Jr Studies in molecular replication. Accts Chem Res. 1994;27:198–203.
-
- Lee DH, Severin K, Yokobayashi Y, Ghadiri MR. Emergence of symbiosis in peptide self-replication through a peptide hypercyclic network. Nature. 1997;390:591–594. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

