An automatic method for CASP9 free modeling structure prediction assessment
- PMID: 21994223
- PMCID: PMC3232368
- DOI: 10.1093/bioinformatics/btr572
An automatic method for CASP9 free modeling structure prediction assessment
Abstract
Motivation: Manual inspection has been applied to and is well accepted for assessing critical assessment of protein structure prediction (CASP) free modeling (FM) category predictions over the years. Such manual assessment requires expertise and significant time investment, yet has the problems of being subjective and unable to differentiate models of similar quality. It is beneficial to incorporate the ideas behind manual inspection to an automatic score system, which could provide objective and reproducible assessment of structure models.
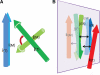
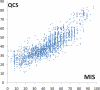
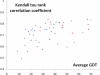
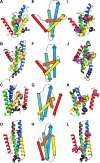
Results: Inspired by our experience in CASP9 FM category assessment, we developed an automatic superimposition independent method named Quality Control Score (QCS) for structure prediction assessment. QCS captures both global and local structural features, with emphasis on global topology. We applied this method to all FM targets from CASP9, and overall the results showed the best agreement with Manual Inspection Scores among automatic prediction assessment methods previously applied in CASPs, such as Global Distance Test Total Score (GDT_TS) and Contact Score (CS). As one of the important components to guide our assessment of CASP9 FM category predictions, this method correlates well with other scoring methods and yet is able to reveal good-quality models that are missed by GDT_TS.
Availability: The script for QCS calculation is available at http://prodata.swmed.edu/QCS/.
Contact: grishin@chop.swmed.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures

References
-
- Aloy P., et al. Predictions without templates: new folds, secondary structure, and contacts in CASP5. Proteins. 2003;53(Suppl. 6):436–456. - PubMed
-
- Ben-David M., et al. Assessment of CASP8 structure predictions for template free targets. Proteins. 2009;77(Suppl. 9):50–65. - PubMed
-
- Jauch R., et al. Assessment of CASP7 structure predictions for template free targets. Proteins. 2007;69(Suppl. 8):57–67. - PubMed
-
- Kinch L.N., et al. CASP5 assessment of fold recognition target predictions. Proteins. 2003;53(Suppl. 6):395–409. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

