Structural characterization of the viral and cRNA panhandle motifs from the infectious salmon anemia virus
- PMID: 21994446
- PMCID: PMC3233167
- DOI: 10.1128/JVI.06250-11
Structural characterization of the viral and cRNA panhandle motifs from the infectious salmon anemia virus
Abstract
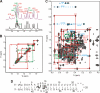
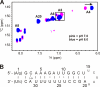
Infectious salmon anemia virus (ISAV) has emerged as a virus of great concern to the aquaculture industry since it can lead to highly contagious and lethal infections in farm-raised salmon populations. While little is known about the transcription/replication cycle of ISAV, initial evidence suggests that it follows molecular mechanisms similar to those found in other orthomyxoviruses, which include the highly pathogenic influenza A (inf A) virus. During the life cycle of orthomyxoviruses, a panhandle structure is formed by the pairing of the conserved 5' and 3' ends of each genomic RNA. This structural motif serves both as a promoter of the viral RNA (vRNA)-dependent RNA polymerase and as a regulatory element in the transcription/replication cycle. As a first step toward characterizing the structure of the ISAV panhandle, here we have determined the secondary structures of the vRNA and the cRNA panhandles on the basis of solution nuclear magnetic resonance (NMR) and thermal melting data. The vRNA panhandle is distinguished by three noncanonical U · G pairs and one U · U pair in two stem helices that are linked by a highly stacked internal loop. For the cRNA panhandle, a contiguous stem helix with a protonated C · A pair near the terminus and tandem downstream U · U pairs was found. The observed noncanonical base pairs and base stacking features of the ISAV RNA panhandle motif provide the first insight into structural features that may govern recognition by the viral RNA polymerase.
Figures




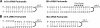
References
-
- Acharya S., et al. 2004. Significant pKa perturbation of nucleobases is an intrinsic property of the sequence context in DNA and RNA. J. Am. Chem. Soc. 126:8674–8681 - PubMed
-
- Batey R. T., Battiste J. L., Williamson J. R. 1995. Preparation of isotopically enriched RNAs for heteronuclear NMR. Methods Enzymol. 261:300–322 - PubMed
-
- Bouchard D. A., Brockway K., Giray C., Keleher W., Merrill P. L. 2001. First report of infectious salmon anemia (ISA) in the United States. Bull. Eur. Assoc. Fish Pathol. 21:86–88
-
- Cai Z., Tinoco I., Jr 1996. Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry 35:6026–6036 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

