Satellite RNA-derived small interfering RNA satsiR-12 targeting the 3' untranslated region of Cucumber mosaic virus triggers viral RNAs for degradation
- PMID: 21994448
- PMCID: PMC3233178
- DOI: 10.1128/JVI.05806-11
Satellite RNA-derived small interfering RNA satsiR-12 targeting the 3' untranslated region of Cucumber mosaic virus triggers viral RNAs for degradation
Abstract
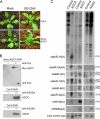
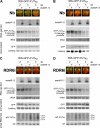
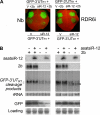
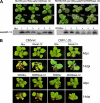
RNA silencing provides protection against RNA viruses by targeting both the helper virus and its satellite RNA (satRNA). Virus-derived small interfering RNAs (vsiRNAs) bound with Argonaute (AGO) proteins are presumed participants in the silencing process. Here, we show that a vsiRNA targeted to virus RNAs triggers the host RNA-dependent RNA polymerase 6 (RDR6)-mediated degradation of viral RNAs. We confirmed that satRNA-derived small interfering RNAs (satsiRNAs) could be associated with different AGO proteins in planta. The most frequently cloned satsiRNA, satsiR-12, was predicted to imperfectly match to Cucumber mosaic virus (CMV) RNAs in the upstream area of the 3' untranslated region (3' UTR). Moreover, an artificial satsiR-12 (asatsiR-12) mediated cleavage of a green fluorescent protein (GFP) sensor construct harboring the satsiR-12 target site. asatsiR-12 also mediated reduction of viral RNAs in 2b-deficient CMV (CMVΔ2b)-infected Nicotiana benthamiana. The reduction was not observed in CMVΔ2b-infected RDR6i plants, in which RDR6 was silenced. Following infection with 2b-containing CMV, the reduction in viral RNAs was not observed in plants of either genotype, indicating that the asatsiR-12-mediated reduction of viral RNAs in the presence of RDR6 was inhibited by the 2b protein. Our results suggest that satsiR-12 targeting the 3' UTR of CMV RNAs triggered RDR6-dependent antiviral silencing. Competition experiments with wild-type CMV RNAs and anti-satsiR-12 mutant RNA1 in the presence of 2b and satRNA demonstrate the inhibitory effect of the 2b protein on the satsiR-12-related degradation of CMV RNAs, revealing a substantial suppressor function of the 2b protein in native CMV infection. Our data provide evidence for the important biological functions of satsiRNAs in homeostatic interactions among the host, virus, and satRNA in the final outcome of viral infection.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

