VPg-primed RNA synthesis of norovirus RNA-dependent RNA polymerases by using a novel cell-based assay
- PMID: 21994457
- PMCID: PMC3233154
- DOI: 10.1128/JVI.06191-11
VPg-primed RNA synthesis of norovirus RNA-dependent RNA polymerases by using a novel cell-based assay
Erratum in
-
Correction for Subba-Reddy et al., "VPg-Primed RNA Synthesis of Norovirus RNA-Dependent RNA Polymerases by Using a Novel Cell-Based Assay".J Virol. 2017 Dec 14;92(1):e01706-17. doi: 10.1128/JVI.01706-17. Print 2018 Jan 1. J Virol. 2017. PMID: 29242253 Free PMC article. No abstract available.
Abstract
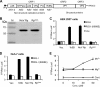
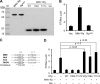
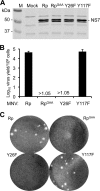
Molecular studies of human noroviruses (NoV) have been hampered by the lack of a permissive cell culture system. We have developed a sensitive and reliable mammalian cell-based assay for the human NoV GII.4 strain RNA-dependent RNA polymerase (RdRp). The assay is based on the finding that RNAs synthesized by transiently expressed RdRp can stimulate retinoic acid-inducible gene I (RIG-I)-dependent reporter luciferase production via the beta interferon promoter. Comparable activities were observed for the murine norovirus (MNV) RdRp. RdRps with mutations at divalent metal ion binding residues did not activate RIG-I signaling. Furthermore, both NoV and MNV RdRp activities were stimulated by the coexpression of their respective VPg proteins, while mutations in the putative site of nucleotide linkage on VPg abolished most of their stimulatory effects. Sequencing of the RNAs linked to VPg revealed that the cellular trans-Golgi network protein 2 (TGOLN2) mRNA was the template for VPg-primed RNA synthesis. Small interfering RNA knockdown of RNase L abolished the enhancement of signaling that occurred in the presence of VPg. Finally, the coexpression of each of the other NoV proteins revealed that p48 (also known as NS1-2) and VP1 enhanced and that VP2 reduced the RdRp activity. The assay should be useful for the dissection of the requirements for NoV RNA synthesis as well as the identification of inhibitors of the NoV RdRp.
Figures
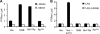





References
-
- Arnold J. J., Ghosh S. K., Cameron C. E. 1999. Poliovirus RNA-dependent RNA polymerase (3Dpol). Divalent cation modulation of primer, template, and nucleotide selection. J. Biol. Chem. 274: 37060–37069 - PubMed
-
- Atmar R. L., Estes M. K. 2006. The epidemiologic and clinical importance of norovirus infection. Gastroenterol. Clin. North Am. 35: 275–290 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

