Quantitative comparison of genetically encoded Ca indicators in cortical pyramidal cells and cerebellar Purkinje cells
- PMID: 21994490
- PMCID: PMC3182323
- DOI: 10.3389/fncel.2011.00018
Quantitative comparison of genetically encoded Ca indicators in cortical pyramidal cells and cerebellar Purkinje cells
Abstract
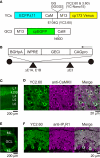
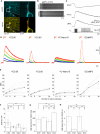
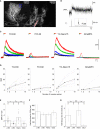
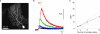
Genetically encoded Ca(2+) indicators (GECIs) are promising tools for cell type-specific and chronic recording of neuronal activity. In the mammalian central nervous system, however, GECIs have been tested almost exclusively in cortical and hippocampal pyramidal cells, and the usefulness of recently developed GECIs has not been systematically examined in other cell types. Here we expressed the latest series of GECIs, yellow cameleon (YC) 2.60, YC3.60, YC-Nano15, and GCaMP3, in mouse cortical pyramidal cells as well as cerebellar Purkinje cells using in utero injection of recombinant adenoviral vectors. We characterized the performance of the GECIs by simultaneous two-photon imaging and whole-cell patch-clamp recording in acute brain slices at 33 ± 2°C. The fluorescent responses of GECIs to action potentials (APs) evoked by somatic current injection or to synaptic stimulation were examined using rapid dendritic imaging. In cortical pyramidal cells, YC2.60 showed the largest responses to single APs, but its decay kinetics were slower than YC3.60 and GCaMP3, while GCaMP3 showed the largest responses to 20 APs evoked at 20 Hz. In cerebellar Purkinje cells, only YC2.60 and YC-Nano15 could reliably report single complex spikes (CSs), and neither showed signal saturation over the entire stimulus range tested (1-10 CSs at 10 Hz). The expression and response of YC2.60 in Purkinje cells remained detectable and comparable for at least over 100 days. These results provide useful information for selecting an optimal GECI depending on the experimental requirements: in cortical pyramidal cells, YC2.60 is suitable for detecting sparse firing of APs, whereas GCaMP3 is suitable for detecting burst firing of APs; in cerebellar Purkinje cells, YC2.60 as well as YC-Nano15 is suitable for detecting CSs.
Keywords: acute brain slice; adenovirus; cerebellar Purkinje cell; cortical pyramidal cell; genetically encoded Ca2+ indicators; patch-clamp recording; two-photon imaging.
Figures
References
-
- Díez-García J., Matsushita S., Mutoh H., Nakai J., Ohkura M., Yokoyama J., Dimitrov D., Knöpfel T. (2005). Activation of cerebellar parallel fibers monitored in transgenic mice expressing a fluorescent Ca2+ indicator protein. Eur. J. Neurosci. 22, 627–63510.1111/j.1460-9568.2005.04250.x - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

