Viroid replication: rolling-circles, enzymes and ribozymes
- PMID: 21994552
- PMCID: PMC3185496
- DOI: 10.3390/v1020317
Viroid replication: rolling-circles, enzymes and ribozymes
Abstract
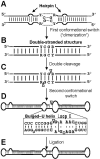
Viroids, due to their small size and lack of protein-coding capacity, must rely essentially on their hosts for replication. Intriguingly, viroids have evolved the ability to replicate in two cellular organella, the nucleus (family Pospiviroidae) and the chloroplast (family Avsunviroidae). Viroid replication proceeds through an RNA-based rolling-circle mechanism with three steps that, with some variations, operate in both polarity strands: i) synthesis of longer-than-unit strands catalyzed by either the nuclear RNA polymerase II or a nuclear-encoded chloroplastic RNA polymerase, in both instances redirected to transcribe RNA templates, ii) cleavage to unit-length, which in the family Avsunviroidae is mediated by hammerhead ribozymes embedded in both polarity strands, while in the family Pospiviroidae the oligomeric RNAs provide the proper conformation but not the catalytic activity, and iii) circularization. The host RNA polymerases, most likely assisted by additional host proteins, start transcription from specific sites, thus implying the existence of viroid promoters. Cleavage and ligation in the family Pospiviroidae is probably catalyzed by an RNase III-like enzyme and an RNA ligase able to circularize the resulting 5' and 3' termini. Whether a chloroplastic RNA ligase mediates circularization in the family Avsunviroidae, or this reaction is autocatalytic, remains an open issue.
Keywords: catalytic RNAs; hammerhead ribozymes; viroids.
Figures


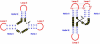
References
-
- Gross HJ, Domdey H, Lossow C, Jank P, Raba M, Alberty H, Sänger HL. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature. 1978;273:203–208. - PubMed
-
- Diener TO. Discovering viroids—a personal perspective. Nat Rev Microbiol. 2003;1:75–80. - PubMed
-
- Tabler M, Tsagris M. Viroids: petite RNA pathogens with distinguished talents. Trends Plant Sci. 2004;9:339–348. - PubMed
-
- Flores R, Hernández C, Martínez de Alba E, Daròs JA, Di Serio F. Viroids and viroid-host interactions. Annu Rev Phytopathol. 2005;43:117–139. - PubMed
LinkOut - more resources
Full Text Sources

