HIV-1 Protease: Structural Perspectives on Drug Resistance
- PMID: 21994585
- PMCID: PMC3185505
- DOI: 10.3390/v1031110
HIV-1 Protease: Structural Perspectives on Drug Resistance
Abstract
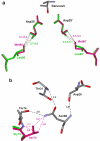
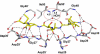
Antiviral inhibitors of HIV-1 protease are a notable success of structure-based drug design and have dramatically improved AIDS therapy. Analysis of the structures and activities of drug resistant protease variants has revealed novel molecular mechanisms of drug resistance and guided the design of tight-binding inhibitors for resistant variants. The plethora of structures reveals distinct molecular mechanisms associated with resistance: mutations that alter the protease interactions with inhibitors or substrates; mutations that alter dimer stability; and distal mutations that transmit changes to the active site. These insights will inform the continuing design of novel antiviral inhibitors targeting resistant strains of HIV.
Keywords: aspartic protease; darunavir; drug resistance; molecular mechanism; protease inhibitors.
Figures

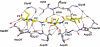
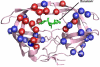
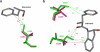
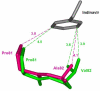


References
-
- Weber IT, Zhang Y, Tozser J. HIV-1 Protease and AIDS Therapy. In: Lendeckel U, Hooper N, editors. Viral Proteases And Antiviral Protease Inhibitor Therapy. in press.
-
- Palella FJ, Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338:853–860. - PubMed
-
- Hammer SM, Squires KE, Hughes MD, Grimes JM, Demeter LM, Currier JS, Eron JJ, Jr, Feinberg JE, Balfour HH, Jr, Deyton LR, Chodakewitz JA, Fischl MA. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N Engl J Med. 1997;337:725–733. - PubMed
-
- Sepkowitz KA. AIDS--the first 20 years. N Eng J Med. 2001;344:1764–1772. - PubMed
-
- Ghosh AK, Chapsal BD, Weber IT, Mitsuya H. Design of HIV protease inhibitors targeting protein backbone: an effective strategy for combating drug resistance. Acc Chem Res. 2008;41:78–86. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

