Satellite RNAs and Satellite Viruses of Plants
- PMID: 21994595
- PMCID: PMC3185516
- DOI: 10.3390/v1031325
Satellite RNAs and Satellite Viruses of Plants
Abstract
The view that satellite RNAs (satRNAs) and satellite viruses are purely molecular parasites of their cognate helper viruses has changed. The molecular mechanisms underlying the synergistic and/or antagonistic interactions among satRNAs/satellite viruses, helper viruses, and host plants are beginning to be comprehended. This review aims to summarize the recent achievements in basic and practical research, with special emphasis on the involvement of RNA silencing mechanisms in the pathogenicity, population dynamics, and, possibly, the origin(s) of these subviral agents. With further research following current trends, the comprehensive understanding of satRNAs and satellite viruses could lead to new insights into the trilateral interactions among host plants, viruses, and satellites.
Keywords: RNA silencing; pathogenicity; replication; satellite RNA; satellite virus.
Figures
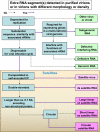
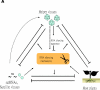

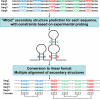
References
-
- Murant AF, Mayo MA. Satellites of plant viruses. Annu Rev Phytopathol. 1982;20:49–70.
-
- Tien P, Wu GS. Satellite RNA for the biocontrol of plant disease. Adv Virus Res. 1991;39:321–39. - PubMed
-
- Garcia-Arenal F, Palukaitis P. Structure and functional relationships of satellite RNAs of cucumber mosaic virus. Curr Top Microbiol Immunol. 1999;239:37–63. - PubMed
-
- Simon AE, Roossinck MJ, Havelda Z. Plant virus satellite and defective interfering RNAs: new paradigms for a new century. Annu Rev Phytopathol. 2004;42:415–37. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

