Initiation of HIV Reverse Transcription
- PMID: 21994608
- PMCID: PMC3185550
- DOI: 10.3390/v2010213
Initiation of HIV Reverse Transcription
Abstract
Reverse transcription of retroviral genomes into double stranded DNA is a key event for viral replication. The very first stage of HIV reverse transcription, the initiation step, involves viral and cellular partners that are selectively packaged into the viral particle, leading to an RNA/protein complex with very specific structural and functional features, some of which being, in the case of HIV-1, linked to particular isolates. Recent understanding of the tight spatio-temporal regulation of reverse transcription and its importance for viral infectivity further points toward reverse transcription and potentially its initiation step as an important drug target.
Keywords: retrovirus; reverse transcriptase; tRNA.
Figures
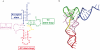

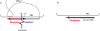
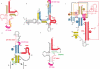


References
-
- Gilboa E, Mitra SW, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. - PubMed
-
- Mölling K, Bolognesi DP, Bauer H, Büsen W, Plassmann HW, Hausen P. Association of viral reverse transcriptase with an enzyme degrading the RNA moiety of RNA-DNA hybrids. Nature New Biol. 1971;234:240–243. - PubMed
-
- Marquet R, Isel C, Ehresmann C, Ehresmann B. tRNAs as primer of reverse transcriptase. Biochimie. 1995;77:113–124. - PubMed
-
- Kleiman L, Halwani R, Javanbakht H. The selective packaging and annealing of primer tRNALys3 in HIV-1. Curr HIV Res. 2004;2:163–175. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

