Jaagsiekte sheep retrovirus biology and oncogenesis
- PMID: 21994634
- PMCID: PMC3185594
- DOI: 10.3390/v2122618
Jaagsiekte sheep retrovirus biology and oncogenesis
Abstract
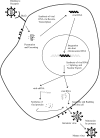
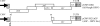
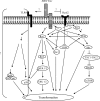
Jaagsiekte sheep retrovirus (JSRV) is the causative agent of a lung cancer in sheep known as ovine pulmonary adenocarcinoma (OPA). The disease has been identified around the world in several breeds of sheep and goats, and JSRV infection typically has a serious impact on affected flocks. In addition, studies on OPA are an excellent model for human lung carcinogenesis. A unique feature of JSRV is that its envelope (Env) protein functions as an oncogene. The JSRV Env-induced transformation or oncogenesis has been studied in a variety of cell systems and in animal models. Moreover, JSRV studies have provided insights into retroviral genomic RNA export/expression mechanisms. JSRV encodes a trans-acting factor (Rej) within the env gene necessary for the synthesis of Gag protein from unspliced viral RNA. This review summarizes research pertaining to JSRV-induced pathogenesis, Env transformation, and other aspects of JSRV biology.
Keywords: Env; JSRV; Jaagsiekte sheep retrovirus; Rej; cancer; ovine pulmonary adenocarcinoma; transformation.
Figures


References
-
- DeMartini JC, Bishop JV, Allen TE, Jassim FA, Sharp JM, de las Heras M, Voelker DR, Carlson JO. Jaagsiekte sheep retrovirus proviral clone JSRV(JS7), derived from the JS7 lung tumor cell line, induces ovine pulmonary carcinoma and is integrated into the surfactant protein A gene. J Virol. 2001;75:4239–4246. - PMC - PubMed
-
- Fan H, Palmarini M, DeMartini JC. Transformation and oncogenesis by jaagsiekte sheep retrovirus. Curr Top Microbiol Immunol. 2003;275:139–177. - PubMed
-
- De las Heras M, Gonzalez L, Sharp JM. Pathology of ovine pulmonary adenocarcinoma. Curr Top Microbiol Immunol. 2003;275:25–54. - PubMed
-
- Sharp JM. Sheep pulmonary adenomatosis: A contagious tumour and its cause. Cancer Surv. 1987;6:73–83. - PubMed

