Complete genomic sequence of bacteriophage felix o1
- PMID: 21994654
- PMCID: PMC3185647
- DOI: 10.3390/v2030710
Complete genomic sequence of bacteriophage felix o1
Abstract
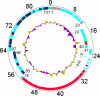
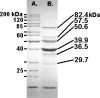
Bacteriophage O1 is a Myoviridae A1 group member used historically for identifying Salmonella. Sequencing revealed a single, linear, 86,155-base-pair genome with 39% average G+C content, 131 open reading frames, and 22 tRNAs. Closest protein homologs occur in Erwinia amylovora phage φEa21-4 and Escherichia coli phage wV8. Proteomic analysis indentified structural proteins: Gp23, Gp36 (major tail protein), Gp49, Gp53, Gp54, Gp55, Gp57, Gp58 (major capsid protein), Gp59, Gp63, Gp64, Gp67, Gp68, Gp69, Gp73, Gp74 and Gp77 (tail fiber). Based on phage-host codon differences, 7 tRNAs could affect translation rate during infection. Introns, holin-lysin cassettes, bacterial toxin homologs and host RNA polymerase-modifying genes were absent.
Keywords: DNA sequence; Felix O1; Myoviridae; Salmonella; bacteriophage; bioinformatics.
Figures

References
-
- Ackermann HW. Salmonella Phages Observed in the Electron Microscope. In: Schatten H, Eisenstark A, editors. Salmonella Methods and Protocols. Humana Press; Totowa, NJ, USA: 2007. pp. 213–234.
-
- Kropinski AM, Sulakvelidze A, Konczy P, Poppe C. Salmonella Phages and Prophages - Genomics and Practical Aspects. In: Schatten H, Eisenstark A, editors. Salmonella Methods and Protocols. Humana Press; Totowa, NJ, USA: 2007. pp. 133–176. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

