Clinical management of HIV drug resistance
- PMID: 21994737
- PMCID: PMC3185705
- DOI: 10.3390/v3040347
Clinical management of HIV drug resistance
Abstract
Combination antiretroviral therapy for HIV-1 infection has resulted in profound reductions in viremia and is associated with marked improvements in morbidity and mortality. Therapy is not curative, however, and prolonged therapy is complicated by drug toxicity and the emergence of drug resistance. Management of clinical drug resistance requires in depth evaluation, and includes extensive history, physical examination and laboratory studies. Appropriate use of resistance testing provides valuable information useful in constructing regimens for treatment-experienced individuals with viremia during therapy. This review outlines the emergence of drug resistance in vivo, and describes clinical evaluation and therapeutic options of the individual with rebound viremia during therapy.
Keywords: HIV; clinical management; drug resistance testing.
Figures



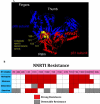


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

