Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood
- PMID: 21994764
- PMCID: PMC3276288
- DOI: 10.1093/hmg/ddr472
Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood
Abstract
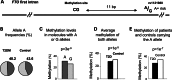
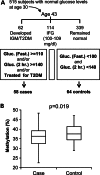
Inter-individual DNA methylation variations were frequently hypothesized to alter individual susceptibility to Type 2 Diabetes Mellitus (T2DM). Sequence-influenced methylations were described in T2DM-associated genomic regions, but evidence for direct, sequence-independent association with disease risk is missing. Here, we explore disease-contributing DNA methylation through a stepwise study design: first, a pool-based, genome-scale screen among 1169 case and control individuals revealed an excess of differentially methylated sites in genomic regions that were previously associated with T2DM through genetic studies. Next, in-depth analyses were performed at selected top-ranking regions. A CpG site in the first intron of the FTO gene showed small (3.35%) but significant (P = 0.000021) hypomethylation of cases relative to controls. The effect was independent of the sequence polymorphism in the region and persists among individuals carrying the sequence-risk alleles. The odds of belonging to the T2DM group increased by 6.1% for every 1% decrease in methylation (OR = 1.061, 95% CI: 1.032-1.090), the odds ratio for decrease of 1 standard deviation of methylation (adjusted to gender) was 1.5856 (95% CI: 1.2824-1.9606) and the sensitivity (area under the curve = 0.638, 95% CI: 0.586-0.690; males = 0.675, females = 0.609) was better than that of the strongest known sequence variant. Furthermore, a prospective study in an independent population cohort revealed significant hypomethylation of young individuals that later progressed to T2DM, relative to the individuals who stayed healthy. Further genomic analysis revealed co-localization with gene enhancers and with binding sites for methylation-sensitive transcriptional regulators. The data showed that low methylation level at the analyzed sites is an early marker of T2DM and suggests a novel mechanism by which early-onset, inter-individual methylation variation at isolated non-promoter genomic sites predisposes to T2DM.
© The Author 2011. Published by Oxford University Press. All rights reserved.
Figures





References
-
- Prokopenko I., McCarthy M.I., Lindgren C.M. Type 2 diabetes: new genes, new understanding. Trends Genet. 2008;24:613–621. doi:10.1016/j.tig.2008.09.004. - DOI - PMC - PubMed
-
- Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P., Zeggini E., Huth C., Aulchenko Y.S., Thorleifsson G., et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat. Genet. 2010;42:579–589. doi:10.1038/ng.609. - DOI - PMC - PubMed
-
- Bjornsson H.T., Fallin M.D., Feinberg A.P. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20:350–358. doi:10.1016/j.tig.2004.06.009. - DOI - PubMed
-
- Portela A., Esteller M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010;28:1057–1068. doi:10.1038/nbt.1685. - DOI - PubMed
-
- Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–727. doi:10.1038/nature09230. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

