HTLV-3/STLV-3 and HTLV-4 viruses: discovery, epidemiology, serology and molecular aspects
- PMID: 21994771
- PMCID: PMC3185789
- DOI: 10.3390/v3071074
HTLV-3/STLV-3 and HTLV-4 viruses: discovery, epidemiology, serology and molecular aspects
Abstract
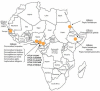
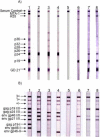
Human T cell leukemia/lymphoma virus Type 1 and 2 (HTLV-1 and HTLV-2), together with their simian counterparts (STLV-1, STLV-2), belong to the Primate T lymphotropic viruses group (PTLV). The high percentage of homologies between HTLV-1 and STLV-1 strains, led to the demonstration that most HTLV-1 subtypes arose from interspecies transmission between monkeys and humans. STLV-3 viruses belong to the third PTLV type and are equally divergent from both HTLV-1 and HTLV-2. They are endemic in several monkey species that live in West, Central and East Africa. In 2005, we, and others reported the discovery of the human homolog (HTLV-3) of STLV-3 in two asymptomatic inhabitants from South Cameroon whose sera exhibited HTLV indeterminate serologies. More recently, two other cases of HTLV-3 infection in persons living in Cameroon were reported suggesting that this virus is not extremely rare in the human population living in Central Africa. Together with STLV-3, these human viral strains belong to the PTLV-3 group. A fourth HTLV type (HTLV-4) was also discovered in the same geographical area. The overall PTLV-3 and PTLV-4 genomic organization is similar to that of HTLV-1 and HTLV-2 with the exception of their long terminal repeats (LTRs) that contain only two 21 bp repeats. As in HTLV-1, HTLV-3 Tax contains a PDZ binding motif while HTLV-4 does not. An antisense transcript was also described in HTLV-3 transfected cells. PTLV-3 molecular clones are now available and will allow scientists to study the viral cycle, the tropism and the possible pathogenicity in vivo. Current studies are also aimed at determining the prevalence, distribution, and modes of transmission of these viruses, as well as their possible association with human diseases. Here we will review the characteristics of these new simian and human retroviruses, whose discovery has opened new avenues of research in the retrovirology field.
Keywords: HTLV-3; HTLV-4; STLV-3; Tax; emerging virus; interspecies transmission; retrovirus; transformation.
Figures



References
-
- Lairmore M, Franchini G. Human T-cell leukemia virus types 1 and 2. In: Wilkins LWA, editor. Field's Virology. 5th ed. Vol. 2. Lippincott Williams & Wilkins; Philadelphia, PA, USA: 2007. pp. 2072–2105.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

