Specialized adaptation of a lactic acid bacterium to the milk environment: the comparative genomics of Streptococcus thermophilus LMD-9
- PMID: 21995282
- PMCID: PMC3231929
- DOI: 10.1186/1475-2859-10-S1-S22
Specialized adaptation of a lactic acid bacterium to the milk environment: the comparative genomics of Streptococcus thermophilus LMD-9
Abstract
Background: Streptococcus thermophilus represents the only species among the streptococci that has "Generally Regarded As Safe" status and that plays an economically important role in the fermentation of yogurt and cheeses. We conducted comparative genome analysis of S. thermophilus LMD-9 to identify unique gene features as well as features that contribute to its adaptation to the dairy environment. In addition, we investigated the transcriptome response of LMD-9 during growth in milk in the presence of Lactobacillus delbrueckii ssp. bulgaricus, a companion culture in yogurt fermentation, and during lytic bacteriophage infection.
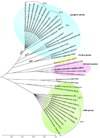
Results: The S. thermophilus LMD-9 genome is comprised of a 1.8 Mbp circular chromosome (39.1% GC; 1,834 predicted open reading frames) and two small cryptic plasmids. Genome comparison with the previously sequenced LMG 18311 and CNRZ1066 strains revealed 114 kb of LMD-9 specific chromosomal region, including genes that encode for histidine biosynthetic pathway, a cell surface proteinase, various host defense mechanisms and a phage remnant. Interestingly, also unique to LMD-9 are genes encoding for a putative mucus-binding protein, a peptide transporter, and exopolysaccharide biosynthetic proteins that have close orthologs in human intestinal microorganisms. LMD-9 harbors a large number of pseudogenes (13% of ORFeome), indicating that like LMG 18311 and CNRZ1066, LMD-9 has also undergone major reductive evolution, with the loss of carbohydrate metabolic genes and virulence genes found in their streptococcal counterparts. Functional genome distribution analysis of ORFeomes among streptococci showed that all three S. thermophilus strains formed a distinct functional cluster, further establishing their specialized adaptation to the nutrient-rich milk niche. An upregulation of CRISPR1 expression in LMD-9 during lytic bacteriophage DT1 infection suggests its protective role against phage invasion. When co-cultured with L. bulgaricus, LMD-9 overexpressed genes involved in amino acid transport and metabolism as well as DNA replication.
Conclusions: The genome of S. thermophilus LMD-9 is shaped by its domestication in the dairy environment, with gene features that conferred rapid growth in milk, stress response mechanisms and host defense systems that are relevant to its industrial applications. The presence of a unique exopolysaccharide gene cluster and cell surface protein orthologs commonly associated with probiotic functionality revealed potential probiotic applications of LMD-9.
Figures



References
-
- Orla-Jensen S. The lactic acid bacteria. Copenhagen: A. F. Host and Son; 1919.
-
- de Vrese M, Stegelmann A, Richter B, Fenselau S, Laue C, Schrezenmeir J. Probiotics--compensation for lactase insufficiency. Am J Clin Nutr. 2001;73:421S–429S. - PubMed
-
- Garcia-Albiach R, Jose M, de Felipe P, Angulo S, Morosini MI, Bravo D, Baquero F, del Campo R. Molecular analysis of yogurt containing Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus in human intestinal microbiota. Am J Clin Nutr. 2008;87:91–96. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

