Multiplexed quantification of nucleic acids with large dynamic range using multivolume digital RT-PCR on a rotational SlipChip tested with HIV and hepatitis C viral load
- PMID: 21995644
- PMCID: PMC3216675
- DOI: 10.1021/ja2060116
Multiplexed quantification of nucleic acids with large dynamic range using multivolume digital RT-PCR on a rotational SlipChip tested with HIV and hepatitis C viral load
Abstract
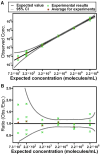
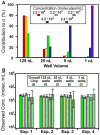
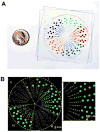
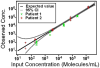
In this paper, we are working toward a problem of great importance to global health: determination of viral HIV and hepatitis C (HCV) loads under point-of-care and resource limited settings. While antiretroviral treatments are becoming widely available, viral load must be evaluated at regular intervals to prevent the spread of drug resistance and requires a quantitative measurement of RNA concentration over a wide dynamic range (from 50 up to 10(6) molecules/mL for HIV and up to 10(8) molecules/mL for HCV). "Digital" single molecule measurements are attractive for quantification, but the dynamic range of such systems is typically limited or requires excessive numbers of compartments. Here we designed and tested two microfluidic rotational SlipChips to perform multivolume digital RT-PCR (MV digital RT-PCR) experiments with large and tunable dynamic range. These designs were characterized using synthetic control RNA and validated with HIV viral RNA and HCV control viral RNA. The first design contained 160 wells of each of four volumes (125 nL, 25 nL, 5 nL, and 1 nL) to achieve a dynamic range of 5.2 × 10(2) to 4.0 × 10(6) molecules/mL at 3-fold resolution. The second design tested the flexibility of this approach, and further expanded it to allow for multiplexing while maintaining a large dynamic range by adding additional wells with volumes of 0.2 nL and 625 nL and dividing the SlipChip into five regions to analyze five samples each at a dynamic range of 1.8 × 10(3) to 1.2 × 10(7) molecules/mL at 3-fold resolution. No evidence of cross-contamination was observed. The multiplexed SlipChip can be used to analyze a single sample at a dynamic range of 1.7 × 10(2) to 2.0 × 10(7) molecules/mL at 3-fold resolution with limit of detection of 40 molecules/mL. HIV viral RNA purified from clinical samples were tested on the SlipChip, and viral load results were self-consistent and in good agreement with results determined using the Roche COBAS AmpliPrep/COBAS TaqMan HIV-1 Test. With further validation, this SlipChip should become useful to precisely quantify viral HIV and HCV RNA for high-performance diagnostics in resource-limited settings. These microfluidic designs should also be valuable for other diagnostic and research applications, including detecting rare cells and rare mutations, prenatal diagnostics, monitoring residual disease, and quantifying copy number variation and gene expression patterns. The theory for the design and analysis of multivolume digital PCR experiments is presented in other work by Kreutz et al.
Figures


References
-
- Marcus JS, Anderson WF, Quake SR. Anal Chem. 2006;78:3084–3089. - PubMed
-
- Stahlberg A, Bengtsson M. Methods. 2010;50:282–288. - PubMed
-
- Grond-Ginsbach C, Hummel M, Wiest T, Horstmann S, Pfleger K, Hergenhahn M, Hollstein M, Mansmann U, Grau AJ, Wagner S. J Neurol. 2008;255:723–731. - PubMed
-
- Kern W, Schoch C, Haferlach T, Schnittger S. Crit Rev Oncol/Hematol. 2005;56:283–309. - PubMed
-
- Schmidt U, Fuessel S, Koch R, Baretton GB, Lohse A, Tomasetti S, Unversucht S, Froehner M, Wirth MP, Meye A. Prostate. 2006;66:1521–1534. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

