Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential
- PMID: 21995752
- PMCID: PMC3212925
- DOI: 10.1186/1472-6750-11-94
Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential
Abstract
Background: To expand on the range of products which can be obtained from lignocellulosic biomass, the lignin component should be utilized as feedstock for value-added chemicals such as substituted aromatics, instead of being incinerated for heat and energy. Enzymes could provide an effective means for lignin depolymerization into products of interest. In this study, soil bacteria were isolated by enrichment on Kraft lignin and evaluated for their ligninolytic potential as a source of novel enzymes for waste lignin valorization.
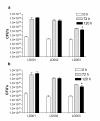
Results: Based on 16S rRNA gene sequencing and phenotypic characterization, the organisms were identified as Pandoraea norimbergensis LD001, Pseudomonas sp LD002 and Bacillus sp LD003. The ligninolytic capability of each of these isolates was assessed by growth on high-molecular weight and low-molecular weight lignin fractions, utilization of lignin-associated aromatic monomers and degradation of ligninolytic indicator dyes. Pandoraea norimbergensis LD001 and Pseudomonas sp. LD002 exhibited best growth on lignin fractions, but limited dye-decolourizing capacity. Bacillus sp. LD003, however, showed least efficient growth on lignin fractions but extensive dye-decolourizing capacity, with a particular preference for the recalcitrant phenothiazine dye class (Azure B, Methylene Blue and Toluidene Blue O).
Conclusions: Bacillus sp. LD003 was selected as a promising source of novel types of ligninolytic enzymes. Our observations suggested that lignin mineralization and depolymerization are separate events which place additional challenges on the screening of ligninolytic microorganisms for specific ligninolytic enzymes.
Figures


References
-
- Martinez AT, Speranza M, Ruiz-Duenas FJ, Ferreira P, Camarero S, Guillen F, Martinez MJ, Gutierrez A, del Rio JC. Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin. Int Microbiol. 2005;8(3):195–204. - PubMed
-
- De los Santos Ramos W, Poznyak T, Chairez I, Cordova RI. Remediation of lignin and its derivatives from pulp and paper industry wastewater by the combination of chemical precipitation and ozonation. Journal of hazardous materials. 2009;169(1-3):428–434. doi: 10.1016/j.jhazmat.2009.03.152. - DOI - PubMed
-
- Stewart D. Lignin as a base material for materials applications: Chemistry, application and economics. Ind Crops Prod. 2008;27:202–207. doi: 10.1016/j.indcrop.2007.07.008. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

