A cis-acting region in the N-methyl-d-aspartate R1 3'-untranslated region interacts with the novel RNA-binding proteins beta subunit of alpha glucosidase II and annexin A2--effect of chronic ethanol exposure in vivo
- PMID: 21995826
- PMCID: PMC3195980
- DOI: 10.1111/j.1460-9568.2011.07857.x
A cis-acting region in the N-methyl-d-aspartate R1 3'-untranslated region interacts with the novel RNA-binding proteins beta subunit of alpha glucosidase II and annexin A2--effect of chronic ethanol exposure in vivo
Abstract
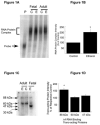
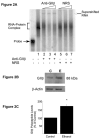
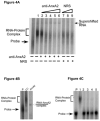
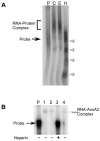
A cis-acting region, Δ4, located in the 3'-untranslated region of N-methyl-d-aspartate R1(NR1) mRNA interacts with several trans-acting proteins present in polysomes purified from fetal cortical neurons. Chronic ethanol exposure of fetal cortical neurons increases Δ4 RNA-protein interactions. This increased interaction is due to an increase in one of the Δ4-binding trans-acting proteins identified as beta subunit of alpha glucosidase II (GIIβ). In this study, we examined whether ethanol-mediated regulation of NR1 mRNA in vivo is similar to that in vitro and whether Δ4-trans interactions are important for ethanol-mediated NR1 mRNA stability. Our data show that polysomal proteins from adult mouse cerebral cortex (CC) formed a complex with Δ4 RNA, suggesting the presence of NR1 mRNA-binding trans-acting proteins in CC polysomes. The intensity of the Δ4 RNA-protein complex was increased with polysomes from chronic ethanol-exposed CC. The Δ4 RNA-protein complex harbored GIIβ and a second trans-acting protein identified as annexin A2 (AnxA2). Ethanol-sensitive GIIβ was upregulated by 70% in ethanol-exposed CC. Heparin, a known binding partner of AnxA2, inhibited Δ4 RNA-protein complex formation. Transient transfection studies using chimeric constructs with and without the Δ4 region revealed that cis-trans interactions are important for ethanol-mediated stability of NR1 mRNA. Furthermore, our data highlight, for the first time, the presence of a binding site on the 3'-untranslated region of NR1 mRNA for AnxA2 and demonstrate the regulation of NR1 mRNA by AnxA2, GIIβ and a third NR1 mRNA-binding protein, which is yet to be identified.
© 2011 The Authors. European Journal of Neuroscience © 2011 Federation of European Neuroscience Societies and Blackwell Publishing Ltd.
Figures


References
-
- Anji A, Kumari M. A novel RNA binding protein that interacts with NMDA R1 mRNA: regulation by ethanol. Eur J Neurosci. 2006;23:2339–2350. - PubMed
-
- Anji A, Shaik KA, Kumari M. Effect of ethanol on lipid-mediated transfection of primary cortical neurons. Ann NY Acad Sci. 2003;993:95–102. discussion 123-104. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

