Reprogramming of mesenchymal stem cells by the synovial sarcoma-associated oncogene SYT-SSX2
- PMID: 21996728
- PMCID: PMC3752676
- DOI: 10.1038/onc.2011.418
Reprogramming of mesenchymal stem cells by the synovial sarcoma-associated oncogene SYT-SSX2
Abstract
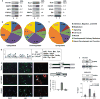
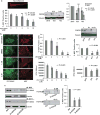
Cell identity is determined by its gene expression programs. The ability of a cell to change its identity and produce cell types outside its lineage is achieved by the activity of transcription controllers capable of reprogramming differentiation gene networks. The synovial sarcoma (SS)-associated protein, SYT-SSX2, reprograms myogenic progenitors and human bone marrow-derived mesenchymal stem cells (BMMSCs) by dictating their commitment to a pro-neural lineage. It fulfills this function by directly targeting an extensive array of neural-specific genes as well as genes of developmental pathway mediators. Concomitantly, the ability of both myoblasts and BMMSCs to differentiate into their normal myogenic and adipogenic lineages was compromised. SS is believed to arise in mesenchymal stem cells where formation of the t(X/18) translocation product, SYT-SSX, constitutes the primary event in the cancer. SYT-SSX is therefore believed to initiate tumorigenesis in its target stem cell. The data presented here allow a glimpse at the initial events that likely occur when SYT-SSX2 is first expressed, and its dominant function in subverting the nuclear program of the stem cell, leading to its aberrant differentiation, as a first step toward transformation. In addition, we identified the fibroblast growth factor receptor gene, Fgfr2, as one occupied and upregulated by SYT-SSX2. Knockdown of FGFR2 in both BMMSCs and SS cells abrogated their growth and attenuated their neural phenotype. These results support the notion that the SYT-SSX2 nuclear function and differentiation effects are conserved throughout sarcoma development and are required for its maintenance beyond the initial phase. They also provide the stem cell regulator, FGFR2, as a promising candidate target for future SS therapy.
Conflict of interest statement
Conflict of interest
The authors declare no conflict of interest.
Figures

References
-
- Chadashvili T, Peterson DA. Cytoarchitecture of fibroblast growth factor receptor 2 (FGFR-2) immunoreactivity in astrocytes of neurogenic and non-neurogenic regions of the young adult and aged rat brain. J Comp Neurol. 2006;498:1–15. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

