Evaluation of model quality predictions in CASP9
- PMID: 21997462
- PMCID: PMC3226935
- DOI: 10.1002/prot.23180
Evaluation of model quality predictions in CASP9
Abstract
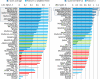
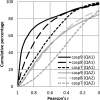
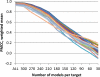
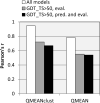
CASP has been assessing the state of the art in the a priori estimation of accuracy of protein structure prediction since 2006. The inclusion of model quality assessment category in CASP contributed to a rapid development of methods in this area. In the last experiment, 46 quality assessment groups tested their approaches to estimate the accuracy of protein models as a whole and/or on a per-residue basis. We assessed the performance of these methods predominantly on the basis of the correlation between the predicted and observed quality of the models on both global and local scales. The ability of the methods to identify the models closest to the best one, to differentiate between good and bad models, and to identify well modeled regions was also analyzed. Our evaluations demonstrate that even though global quality assessment methods seem to approach perfection point (weighted average per-target Pearson's correlation coefficients are as high as 0.97 for the best groups), there is still room for improvement. First, all top-performing methods use consensus approaches to generate quality estimates, and this strategy has its own limitations. Second, the methods that are based on the analysis of individual models lag far behind clustering techniques and need a boost in performance. The methods for estimating per-residue accuracy of models are less accurate than global quality assessment methods, with an average weighted per-model correlation coefficient in the range of 0.63-0.72 for the best 10 groups.
Copyright © 2011 Wiley-Liss, Inc.
Figures




References
-
- Schwede T, Sali A, Honig B, Levitt M, Berman HM, Jones D, Brenner SE, Burley SK, Das R, Dokholyan NV, Dunbrack RL, Jr., Fidelis K, Fiser A, Godzik A, Huang YJ, Humblet C, Jacobson MP, Joachimiak A, Krystek SR, Jr., Kortemme T, Kryshtafovych A, Montelione GT, Moult J, Murray D, Sanchez R, Sosnick TR, Standley DM, Stouch T, Vajda S, Vasquez M, Westbrook JD, Wilson IA. Outcome of a Workshop on Applications of Protein Models in Biomedical Research. Structure. 2009;17(2):151–159. - PMC - PubMed
-
- Moult J. Comparative modeling in structural genomics. Structure. 2008;16(1):14–16. - PubMed
-
- Tramontano A. The role of molecular modelling in biomedical research. FEBS Lett. 2006;580(12):2928–2934. - PubMed
-
- Haider SM, Patel JS, Poojari CS, Neidle S. Molecular modeling on inhibitor complexes and active-site dynamics of cytochrome P450 C17, a target for prostate cancer therapy. J Mol Biol. 2010;400(5):1078–1098. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

