CASP9 assessment of free modeling target predictions
- PMID: 21997521
- PMCID: PMC3226891
- DOI: 10.1002/prot.23181
CASP9 assessment of free modeling target predictions
Abstract
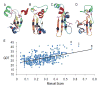
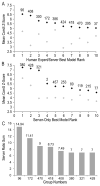
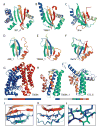
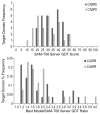
We present an overview of the ninth round of Critical Assessment of Protein Structure Prediction (CASP9) "Template free modeling" category (FM). Prediction models were evaluated using a combination of established structural and sequence comparison measures and a novel automated method designed to mimic manual inspection by capturing both global and local structural features. These scores were compared to those assigned manually over a diverse subset of target domains. Scores were combined to compare overall performance of participating groups and to estimate rank significance. Moreover, we discuss a few examples of free modeling targets to highlight the progress and bottlenecks of current prediction methods. Notably, a server prediction model for a single target (T0581) improved significantly over the closest structure template (44% GDT increase). This accomplishment represents the "winner" of the CASP9 FM category. A number of human expert groups submitted slight variations of this model, highlighting a trend for human experts to act as "meta predictors" by correctly selecting among models produced by the top-performing automated servers. The details of evaluation are available at http://prodata.swmed.edu/CASP9/ .
Copyright © 2011 Wiley-Liss, Inc.
Figures



References
-
- Simons KT, Bonneau R, Ruczinski I, Baker D. Ab initio protein structure prediction of CASP III targets using ROSETTA. Proteins. 1999;(Suppl 3):171–176. - PubMed
-
- Das R, Qian B, Raman S, Vernon R, Thompson J, Bradley P, Khare S, Tyka MD, Bhat D, Chivian D, Kim DE, Sheffler WH, Malmstrom L, Wollacott AM, Wang C, Andre I, Baker D. Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home. Proteins. 2007;69 (Suppl 8):118–128. - PubMed
-
- Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69 (Suppl 8):108–117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

