A-MYB (MYBL1) stimulates murine testis-specific Ldhc expression via the cAMP-responsive element (CRE) site
- PMID: 21998171
- PMCID: PMC3290662
- DOI: 10.1095/biolreprod.111.095661
A-MYB (MYBL1) stimulates murine testis-specific Ldhc expression via the cAMP-responsive element (CRE) site
Abstract
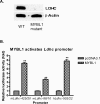
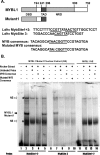
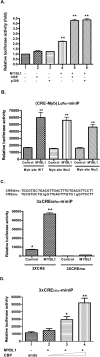
Generally, knowledge of the mechanism regulating gene expression in primary spermatocytes is incomplete. We have used the lactate dehydrogenase gene (Ldhc) as a model to explore these mechanisms during spermatogenesis. Its 100-bp core promoter contained two essential elements common to many genes, a GC box and a CRE site. Here we report results that support a model in which transcription factor MYBL1 acts as a coactivator directing tissue-specific expression via the CRE cis element. We hypothesize that this is a common mechanism involving activation of multiple genes in the primary spermatocyte. MYBL1 is expressed predominantly as a tissue-specific transcription factor in spermatocytes and breast epithelial cells. Our finding that LDHC expression is lost in 21-day testes of MYBL1 mutant mice supports our hypothesis. In the GC1-spg germ cell line exogenous MYBL1 induces activity 4- to 8-fold, although extracts from these cells do not show MYBL1 binding activity for the Myb consensus sequences in the Ldhc promoter by EMSA. Rather, MYBL1 stimulates expression from a synthetic promoter containing only CRE elements, suggesting MYBL1 activates the promoter by interacting with protein that binds to a CRE element. Mutation of three Myb sites does not affect Ldhc promoter activity significantly (P > 0.05). CREB-binding protein (CBP) is a coactivator that interacts with CRE-binding protein CREB. We show that the transactivation domain (TAD) in MYBL1 interacts with the KIX domain in CBP, and the TAD domain and DNA binding domain in MYBL1 each interact with the CREB N-terminal domain. MYBL1 also stimulated expression from testis-specific genes Pgk2 (phosphoglycerate kinase 2) and Pdha2 (pyruvate dehydrogenase alpha 2) promoters, each of which contains CRE promoter elements and is expressed in primary spermatocytes. We propose that MYBL1 directs germ cell-specific activation via the CRE site of certain genes that are activated specifically in the primary spermatocyte, although other, more indirect effects of MYBL1 remain a possible explanation for our results.
Figures




References
-
- Naz RK, Rajesh P. Novel testis/sperm-specific contraceptive targets identified using gene knockout studies. Front Biosci 2005; 10: 2430 2446 - PubMed
-
- Naz RK. Alexis Engle R. Gene knockouts that affect male fertility: novel targets for contraception. Front Biosci 2009; 14: 3994 4007 - PubMed
-
- MacLean J, II, Wilkinson M. Gene regulation in spermatogenesis. Curr Topics Dev Biol 2005; 71: 131 197 - PubMed
-
- Macho B, Brancorsini S, Fimia GM, Setou M, Hirokawa N, Sassone-Corsi P. CREM-dependent transcription in male germ cells controlled by a kinesin. Science 2002; 298: 2388 2390 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

