Defining pheromone-receptor signaling in Candida albicans and related asexual Candida species
- PMID: 21998194
- PMCID: PMC3237633
- DOI: 10.1091/mbc.E11-09-0749
Defining pheromone-receptor signaling in Candida albicans and related asexual Candida species
Erratum in
- Mol Biol Cell. 2012 Mar;23(6):1140
Abstract
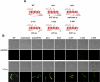
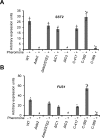
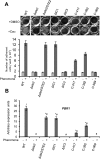
Candida albicans is an important human fungal pathogen in which sexual reproduction is under the control of the novel white-opaque switch. Opaque cells are the mating-competent form, whereas white cells do not mate but can still respond to pheromones, resulting in biofilm formation. In this study, we first define the domains of the α-pheromone receptor Ste2 that are necessary for signaling in both white and opaque forms. Both cell states require the IC loop 3 (IC3) and the C-terminal tail of Ste2 for the cellular response, whereas the first IC loop (IC1) of Ste2 is dispensable for signaling. To also address pheromone-receptor interactions in related species, including apparently asexual Candida species, Ste2 orthologues were heterologously expressed in Candida albicans. Ste2 receptors from multiple Candida clade species were functional when expressed in C. albicans, whereas the Ste2 receptor of Candida lusitaniae was nonfunctional. Significantly, however, expression of a chimeric C. lusitaniae Ste2 receptor containing the C-terminal tail of Ste2 from C. albicans generated a productive response to C. lusitaniae pheromone. This system has allowed us to characterize pheromones from multiple Candida species and indicates that functional pheromone-receptor couples exist in fungal species that have yet to be shown to undergo sexual mating.
Figures
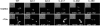





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

