c-Fos activates and physically interacts with specific enzymes of the pathway of synthesis of polyphosphoinositides
- PMID: 21998197
- PMCID: PMC3237616
- DOI: 10.1091/mbc.E11-03-0259
c-Fos activates and physically interacts with specific enzymes of the pathway of synthesis of polyphosphoinositides
Abstract
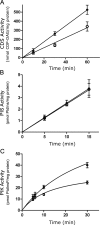
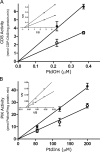
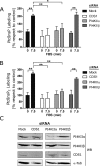
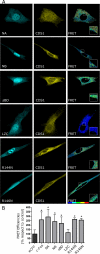
The oncoprotein c-Fos is a well-recognized AP-1 transcription factor. In addition, this protein associates with the endoplasmic reticulum and activates the synthesis of phospholipids. However, the mechanism by which c-Fos stimulates the synthesis of phospholipids in general and the specific lipid pathways activated are unknown. Here we show that induction of quiescent cells to reenter growth promotes an increase in the labeling of polyphosphoinositides that depends on the expression of c-Fos. We also investigated whether stimulation by c-Fos of the synthesis of phosphatidylinositol and its phosphorylated derivatives depends on the activation of enzymes of the phosphatidylinositolphosphate biosynthetic pathway. We found that c-Fos activates CDP-diacylglycerol synthase and phosphatidylinositol (PtdIns) 4-kinase II α in vitro, whereas no activation of phosphatidylinositol synthase or of PtdIns 4-kinase II β was observed. Both coimmunoprecipitation and fluorescence resonance energy transfer experiments consistently showed a physical interaction between the N-terminal domain of c-Fos and the enzymes it activates.
Figures





References
-
- Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. - PubMed
-
- Antonsson B. Phosphatidylinositol synthase from mammalian tissues. Biochim Biophys Acta. 1997;1348:179–186. - PubMed
-
- Balla A, Tuymetova G, Barshishat M, Geiszt M, Balla T. Characterization of type II phosphatidylinositol 4-kinase isoforms reveals association of the enzymes with endosomal vesicular compartments. J Biol Chem. 2002;277:20041–20050. - PubMed
-
- Balla A, Balla T. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol. 2006;16:351–361. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

