Topological constraints impair RNA polymerase II transcription and causes instability of plasmid-borne convergent genes
- PMID: 21998294
- PMCID: PMC3273821
- DOI: 10.1093/nar/gkr840
Topological constraints impair RNA polymerase II transcription and causes instability of plasmid-borne convergent genes
Abstract
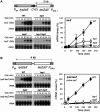
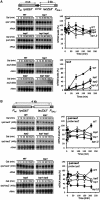
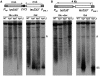
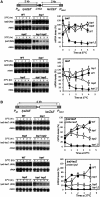
Despite the theoretical bases for the association of topoisomerases and supercoiling changes with transcription and replication, our knowledge of the impact of topological constraints on transcription and replication is incomplete. Although mutation of topoisomerases affects expression and stability of the rDNA region it is not clear whether the same is the case for RNAPII transcription and genome integrity in other regions. We developed new assays in which two convergent RNAPII-driven genes are transcribed simultaneously. Plasmid-based systems were constructed with and without a transcription terminator between the two convergent transcription units, so that the impact of transcription interference could also be evaluated. Using these assays we show that Topos I and II play roles in RNAPII transcription in vivo and reduce the stability of RNAPII-transcribed genes in Saccharomyces cerevisiae. Supercoiling accumulation in convergent transcription units impairs RNAPII transcription in top1Δ strains, but Topo II is also required for efficient transcription independent of Topo I and of detectable supercoiling accumulation. Our work shows that topological constraints negatively affect RNAPII transcription and genetic integrity, and provides an assay to study gene regulation by transcription interference.
Figures



References
-
- Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. - PubMed
-
- Crooke E, Hwang DS, Skarstad K, Thony B, Kornberg A. E. coli minichromosome replication: regulation of initiation at oriC. Res. Microbiol. 1991;142:127–130. - PubMed
-
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell. Biol. 2002;3:430–440. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

