Contributions of the peroxisome and β-oxidation cycle to biotin synthesis in fungi
- PMID: 21998305
- PMCID: PMC3234907
- DOI: 10.1074/jbc.M111.279687
Contributions of the peroxisome and β-oxidation cycle to biotin synthesis in fungi
Abstract
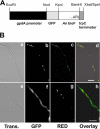
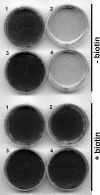
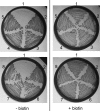
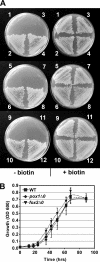
The first step in the synthesis of the bicyclic rings of D-biotin is mediated by 8-amino-7-oxononanoate (AON) synthase, which catalyzes the decarboxylative condensation of l-alanine and pimelate thioester. We found that the Aspergillus nidulans AON synthase, encoded by the bioF gene, is a peroxisomal enzyme with a type 1 peroxisomal targeting sequence (PTS1). Localization of AON to the peroxisome was essential for biotin synthesis because expression of a cytosolic AON variant or deletion of pexE, encoding the PTS1 receptor, rendered A. nidulans a biotin auxotroph. AON synthases with PTS1 are found throughout the fungal kingdom, in ascomycetes, basidiomycetes, and members of basal fungal lineages but not in representatives of the Saccharomyces species complex, including Saccharomyces cerevisiae. A. nidulans mutants defective in the peroxisomal acyl-CoA oxidase AoxA or the multifunctional protein FoxA showed a strong decrease in colonial growth rate in biotin-deficient medium, whereas partial growth recovery occurred with pimelic acid supplementation. These results indicate that pimeloyl-CoA is the in vivo substrate of AON synthase and that it is generated in the peroxisome via the β-oxidation cycle in A. nidulans and probably in a broad range of fungi. However, the β-oxidation cycle is not essential for biotin synthesis in S. cerevisiae or Escherichia coli. These results suggest that alternative pathways for synthesis of the pimelate intermediate exist in bacteria and eukaryotes and that Saccharomyces species use a pathway different from that used by the majority of fungi.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

