The Escherichia coli replisome is inherently DNA damage tolerant
- PMID: 21998391
- PMCID: PMC3593629
- DOI: 10.1126/science.1209111
The Escherichia coli replisome is inherently DNA damage tolerant
Abstract
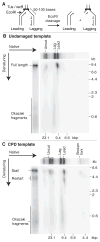
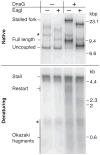
The Escherichia coli DNA replication machinery must frequently overcome template lesions under normal growth conditions. Yet, the outcome of a collision between the replisome and a leading-strand template lesion remains poorly understood. Here, we demonstrate that a single, site-specific, cyclobutane pyrimidine dimer leading-strand template lesion provides only a transient block to fork progression in vitro. The replisome remains stably associated with the fork after collision with the lesion. Leading-strand synthesis is then reinitiated downstream of the damage in a reaction that is dependent on the primase, DnaG, but independent of any of the known replication-restart proteins. These observations reveal that the replisome can tolerate leading-strand template lesions without dissociating by synthesizing the leading strand discontinuously.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

