NMR detection of structures in the HIV-1 5'-leader RNA that regulate genome packaging
- PMID: 21998393
- PMCID: PMC3335204
- DOI: 10.1126/science.1210460
NMR detection of structures in the HIV-1 5'-leader RNA that regulate genome packaging
Abstract
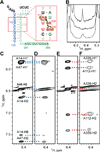
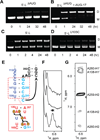
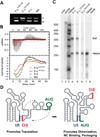
The 5'-leader of the HIV-1 genome regulates multiple functions during viral replication via mechanisms that have yet to be established. We developed a nuclear magnetic resonance approach that enabled direct detection of structural elements within the intact leader (712-nucleotide dimer) that are critical for genome packaging. Residues spanning the gag start codon (AUG) form a hairpin in the monomeric leader and base pair with residues of the unique-5' region (U5) in the dimer. U5:AUG formation promotes dimerization by displacing and exposing a dimer-promoting hairpin and enhances binding by the nucleocapsid (NC) protein, which is the cognate domain of the viral Gag polyprotein that directs packaging. Our findings support a packaging mechanism in which translation, dimerization, NC binding, and packaging are regulated by a common RNA structural switch.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

