The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine
- PMID: 21998396
- PMCID: PMC3321924
- DOI: 10.1126/science.1209791
The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine
Abstract
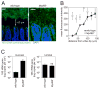
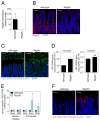
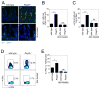
The mammalian intestine is home to ~100 trillion bacteria that perform important metabolic functions for their hosts. The proximity of vast numbers of bacteria to host intestinal tissues raises the question of how symbiotic host-bacterial relationships are maintained without eliciting potentially harmful immune responses. Here, we show that RegIIIγ, a secreted antibacterial lectin, is essential for maintaining a ~50-micrometer zone that physically separates the microbiota from the small intestinal epithelial surface. Loss of host-bacterial segregation in RegIIIγ(-/-) mice was coupled to increased bacterial colonization of the intestinal epithelial surface and enhanced activation of intestinal adaptive immune responses by the microbiota. Together, our findings reveal that RegIIIγ is a fundamental immune mechanism that promotes host-bacterial mutualism by regulating the spatial relationships between microbiota and host.
Figures

Comment in
-
Microbiology. Keeping bacteria at a distance.Science. 2011 Oct 14;334(6053):182-3. doi: 10.1126/science.1213909. Science. 2011. PMID: 21998374 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

