Change in allosteric network affects binding affinities of PDZ domains: analysis through perturbation response scanning
- PMID: 21998559
- PMCID: PMC3188487
- DOI: 10.1371/journal.pcbi.1002154
Change in allosteric network affects binding affinities of PDZ domains: analysis through perturbation response scanning
Abstract
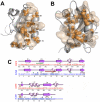
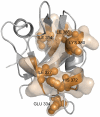
The allosteric mechanism plays a key role in cellular functions of several PDZ domain proteins (PDZs) and is directly linked to pharmaceutical applications; however, it is a challenge to elaborate the nature and extent of these allosteric interactions. One solution to this problem is to explore the dynamics of PDZs, which may provide insights about how intramolecular communication occurs within a single domain. Here, we develop an advancement of perturbation response scanning (PRS) that couples elastic network models with linear response theory (LRT) to predict key residues in allosteric transitions of the two most studied PDZs (PSD-95 PDZ3 domain and hPTP1E PDZ2 domain). With PRS, we first identify the residues that give the highest mean square fluctuation response upon perturbing the binding sites. Strikingly, we observe that the residues with the highest mean square fluctuation response agree with experimentally determined residues involved in allosteric transitions. Second, we construct the allosteric pathways by linking the residues giving the same directional response upon perturbation of the binding sites. The predicted intramolecular communication pathways reveal that PSD-95 and hPTP1E have different pathways through the dynamic coupling of different residue pairs. Moreover, our analysis provides a molecular understanding of experimentally observed hidden allostery of PSD-95. We show that removing the distal third alpha helix from the binding site alters the allosteric pathway and decreases the binding affinity. Overall, these results indicate that (i) dynamics plays a key role in allosteric regulations of PDZs, (ii) the local changes in the residue interactions can lead to significant changes in the dynamics of allosteric regulations, and (iii) this might be the mechanism that each PDZ uses to tailor their binding specificities regulation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Goodey NM, Benkovic SJ. Allosteric regulation and catalysis emerge via a common route. Nat Chem Biol. 2008;4:474–482. - PubMed
-
- Monod J, Wyman J, Changeux JP. On the Nature of Allosteric Transitions: a Plausible Model. J Mol Biol. 1965;12:88–118. - PubMed
-
- Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. - PubMed
-
- Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

