High-precision, in vitro validation of the sequestration mechanism for generating ultrasensitive dose-response curves in regulatory networks
- PMID: 21998566
- PMCID: PMC3188500
- DOI: 10.1371/journal.pcbi.1002171
High-precision, in vitro validation of the sequestration mechanism for generating ultrasensitive dose-response curves in regulatory networks
Abstract
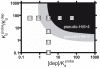
Our ability to recreate complex biochemical mechanisms in designed, artificial systems provides a stringent test of our understanding of these mechanisms and opens the door to their exploitation in artificial biotechnologies. Motivated by this philosophy, here we have recapitulated in vitro the "target sequestration" mechanism used by nature to improve the sensitivity (the steepness of the input/output curve) of many regulatory cascades. Specifically, we have employed molecular beacons, a commonly employed optical DNA sensor, to recreate the sequestration mechanism and performed an exhaustive, quantitative study of its key determinants (e.g., the relative concentrations and affinities of probe and depletant). We show that, using sequestration, we can narrow the pseudo-linear range of a traditional molecular beacon from 81-fold (i.e., the transition from 10% to 90% target occupancy spans an 81-fold change in target concentration) to just 1.5-fold. This narrowing of the dynamic range improves the sensitivity of molecular beacons to that equivalent of an oligomeric, allosteric receptor with a Hill coefficient greater than 9. Following this we have adapted the sequestration mechanism to steepen the binding-site occupancy curve of a common transcription factor by an order of magnitude over the sensitivity observed in the absence of sequestration. Given the success with which the sequestration mechanism has been employed by nature, we believe that this strategy could dramatically improve the performance of synthetic biological systems and artificial biosensors.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

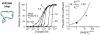
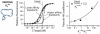
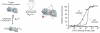
References
-
- Guet CC, Elowitz MB, Hsing W, Leibler S. Combinatorial synthesis of genetic networks. Science. 2002;296:1466–1470. - PubMed
-
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in escherichia coli. Nature. 2000;403:339–342. - PubMed
-
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

