Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos
- PMID: 21998593
- PMCID: PMC3188537
- DOI: 10.1371/journal.pgen.1002279
Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos
Abstract
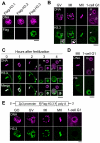
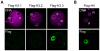
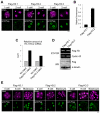
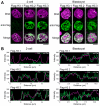
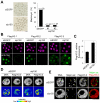
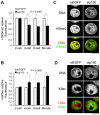
Upon fertilization, reprogramming of gene expression is required for embryo development. This step is marked by DNA demethylation and changes in histone variant composition. However, little is known about the molecular mechanisms causing these changes and their impact on histone modifications. We examined the global deposition of the DNA replication-dependent histone H3.1 and H3.2 variants and the DNA replication-independent H3.3 variant after fertilization in mice. We showed that H3.3, a euchromatic marker of gene activity, transiently disappears from the maternal genome, suggesting erasure of the oocyte-specific modifications carried by H3.3. After fertilization, H3.2 is incorporated into the transcriptionally silent heterochromatin, whereas H3.1 and H3.3 occupy unusual heterochromatic and euchromatin locations, respectively. After the two-cell stage, H3.1 and H3.3 variants resume their usual respective locations on heterochromatin and euchromatin. Preventing the incorporation of H3.1 and H3.2 by knockdown of the histone chaperone CAF-1 induces a reciprocal increase in H3.3 deposition and impairs heterochromatin formation. We propose that the deposition of different H3 variants influences the functional organization of chromatin. Taken together, these findings suggest that dynamic changes in the deposition of H3 variants are critical for chromatin reorganization during epigenetic reprogramming.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. - PubMed
-
- Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Grunstein M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. - PubMed
-
- Jeppesen P. Histone acetylation: a possible mechanism for the inheritance of cell memory at mitosis. Bioessays. 1997;19:67–74. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

