Fluorescent labeling of antibody fragments using split GFP
- PMID: 21998685
- PMCID: PMC3187779
- DOI: 10.1371/journal.pone.0025727
Fluorescent labeling of antibody fragments using split GFP
Abstract
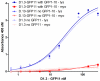
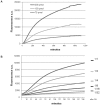
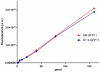
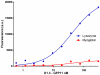
Antibody fragments are easily isolated from in vitro selection systems, such as phage and yeast display. Lacking the Fc portion of the antibody, they are usually labeled using small peptide tags recognized by antibodies. In this paper we present an efficient method to fluorescently label single chain Fvs (scFvs) using the split green fluorescent protein (GFP) system. A 13 amino acid tag, derived from the last beta strand of GFP (termed GFP11), is fused to the C terminus of the scFv. This tag has been engineered to be non-perturbing, and we were able to show that it exerted no effect on scFv expression or functionality when compared to a scFv without the GFP11 tag. Effective functional fluorescent labeling is demonstrated in a number of different assays, including fluorescence linked immunosorbant assays, flow cytometry and yeast display. Furthermore, we were able to show that this split GFP system can be used to determine the concentration of scFv in crude samples, as well an estimate of antibody affinity, without the need for antibody purification. We anticipate this system will be of widespread interest in antibody engineering and in vitro display systems.
Conflict of interest statement
Figures

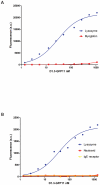
References
-
- Bradbury AR, Marks JD. Antibodies from phage antibody libraries. J Immunol Methods. 2004;290:29–49. - PubMed
-
- Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, et al. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222:581–597. - PubMed
-
- Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

