The mitochondrial genomes of the early land plants Treubia lacunosa and Anomodon rugelii: dynamic and conservative evolution
- PMID: 21998706
- PMCID: PMC3187804
- DOI: 10.1371/journal.pone.0025836
The mitochondrial genomes of the early land plants Treubia lacunosa and Anomodon rugelii: dynamic and conservative evolution
Abstract
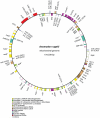
Early land plant mitochondrial genomes captured important changes of mitochondrial genome evolution when plants colonized land. The chondromes of seed plants show several derived characteristics, e.g., large genome size variation, rapid intra-genomic rearrangement, abundant introns, and highly variable levels of RNA editing. On the other hand, the chondromes of charophytic algae are still largely ancestral in these aspects, resembling those of early eukaryotes. When the transition happened has been a long-standing question in studies of mitochondrial genome evolution. Here we report complete mitochondrial genome sequences from an early-diverging liverwort, Treubia lacunosa, and a late-evolving moss, Anomodon rugelii. The two genomes, 151,983 and 104,239 base pairs in size respectively, contain standard sets of protein coding genes for respiration and protein synthesis, as well as nearly full sets of rRNA and tRNA genes found in the chondromes of the liverworts Marchantia polymorpha and Pleurozia purpurea and the moss Physcomitrella patens. The gene orders of these two chondromes are identical to those of the other liverworts and moss. Their intron contents, with all cis-spliced group I or group II introns, are also similar to those in the previously sequenced liverwort and moss chondromes. These five chondromes plus the two from the hornworts Phaeoceros laevis and Megaceros aenigmaticus for the first time allowed comprehensive comparative analyses of structure and organization of mitochondrial genomes both within and across the three major lineages of bryophytes. These analyses led to the conclusion that the mitochondrial genome experienced dynamic evolution in genome size, gene content, intron acquisition, gene order, and RNA editing during the origins of land plants and their major clades. However, evolution of this organellar genome has remained rather conservative since the origin and initial radiation of early land plants, except within vascular plants.
Conflict of interest statement
Figures


References
-
- Schuster W, Brennicke A. The plant mitochondrial genome - physical structure, information content, RNA editing, and gene migration to the nucleus. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:61–78.
-
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. - PubMed
-
- Knoop V. The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr Genet. 2004;46:123–139. - PubMed
-
- Knoop V, Volkmar U, Hecht J, Grewe F. Mitochondrial genome evolution in the plant lineage. In: Kempken F, editor. Plant Mitochondria. Dordrecht: Springer; 2010. pp. 3–29.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

