A human multi-epitope recombinant vaccinia virus as a universal T cell vaccine candidate against influenza virus
- PMID: 21998725
- PMCID: PMC3187825
- DOI: 10.1371/journal.pone.0025938
A human multi-epitope recombinant vaccinia virus as a universal T cell vaccine candidate against influenza virus
Abstract
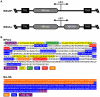
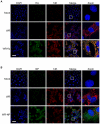
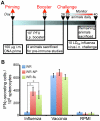
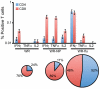
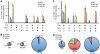
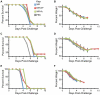
There is a need to develop a universal vaccine against influenza virus infection to avoid developing new formulations of a seasonal vaccine each year. Many of the vaccine strategies for a universal vaccine target strain-conserved influenza virus proteins, such as the matrix, polymerase, and nucleoproteins, rather than the surface hemagglutinin and neuraminidase proteins. In addition, non-disease-causing viral vectors are a popular choice as a delivery system for the influenza virus antigens. As a proof-of-concept, we have designed a novel influenza virus immunogen based on the NP backbone containing human T cell epitopes for M1, NS1, NP, PB1 and PA proteins (referred as NPmix) as well as a construct containing the conserved regions of influenza virus neuraminidase (N-terminal) and hemagglutinin (C-terminal) (referred as NA-HA). DNA vectors and vaccinia virus recombinants expressing NPmix (WR-NP) or both NPmix plus NA-HA (WR-flu) in the cytosol were tested in a heterologous DNA-prime/vaccinia virus-boost vaccine regimen in mice. We observed an increase in the number of influenza virus-specific IFNγ-secreting splenocytes, composed of populations marked by CD4(+) and CD8(+) T cells producing IFNγ or TNFα. Upon challenge with influenza virus, the vaccinated mice exhibited decreased viral load in the lungs and a delay in mortality. These findings suggest that DNA prime/poxvirus boost with human multi-epitope recombinant influenza virus proteins is a valid approach for a general T-cell vaccine to protect against influenza virus infection.
Conflict of interest statement
Figures

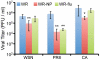
References
-
- Regoes RR, Bonhoeffer S. Emergence of Drug-Resistant Influenza Virus: Population Dynamical Considerations. Science. 2006;312:389–391. - PubMed
-
- Gibbs MJ, Armstrong JS, Gibbs AJ. Recombination in the Hemagglutinin Gene of the 1918 “Spanish Flu”. Science. 2001;293:1842–1845. - PubMed
-
- Li KS, Guan Y, Wang J, Smith GJD, Xu KM, et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. - PubMed
-
- Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, et al. Influenza-associated hospitalizations in the United States. JAMA. 2004;292:1333–1340. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

