Generating a knockdown transgene against Drosophila heterochromatic Tim17b gene encoding mitochondrial translocase subunit
- PMID: 21998726
- PMCID: PMC3188573
- DOI: 10.1371/journal.pone.0025945
Generating a knockdown transgene against Drosophila heterochromatic Tim17b gene encoding mitochondrial translocase subunit
Abstract
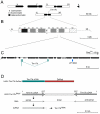
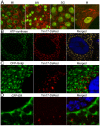
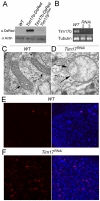
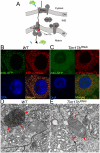
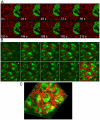
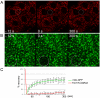
Heterochromatic regions of eukaryotic genomes contain multiple functional elements involved in chromosomal dynamics, as well as multiple housekeeping genes. Cytological and molecular peculiarities of heterochromatic loci complicate genetic studies based on standard approaches developed using euchromatic genes. Here, we report the development of an RNAi-based knockdown transgenic construct and red fluorescent reporter transgene for a small gene, Tim17b, which localizes in constitutive heterochromatin of Drosophila melanogaster third chromosome and encodes a mitochondrial translocase subunit. We demonstrate that Tim17b protein is required strictly for protein delivery to mitochondrial matrix. Knockdown of Tim17b completely disrupts functions of the mitochondrial translocase complex. Using fluorescent recovery after photobleaching assay, we show that Tim17b protein has a very stable localization in the membranes of the mitochondrial network and that its exchange rate is close to zero when compared with soluble proteins of mitochondrial matrix. These results confirm that we have developed comprehensive tools to study functions of heterochromatic Tim17b gene.
Conflict of interest statement
Figures
Similar articles
-
Mitochondrial protein import regulates cytosolic protein homeostasis and neuronal integrity.Autophagy. 2018;14(8):1293-1309. doi: 10.1080/15548627.2018.1474991. Epub 2018 Jul 21. Autophagy. 2018. PMID: 29909722 Free PMC article.
-
Drosophila oncogene Gas41 is an RNA interference modulator that intersects heterochromatin and the small interfering RNA pathway.FEBS J. 2015 Jan;282(1):153-73. doi: 10.1111/febs.13115. Epub 2014 Nov 17. FEBS J. 2015. PMID: 25323651
-
Heterochromatic deposition of centromeric histone H3-like proteins.Proc Natl Acad Sci U S A. 2000 Jan 18;97(2):716-21. doi: 10.1073/pnas.97.2.716. Proc Natl Acad Sci U S A. 2000. PMID: 10639145 Free PMC article.
-
Drosophila melanogaster as a model for studying protein-encoding genes that are resident in constitutive heterochromatin.Heredity (Edinb). 2007 Jan;98(1):3-12. doi: 10.1038/sj.hdy.6800877. Epub 2006 Nov 1. Heredity (Edinb). 2007. PMID: 17080025 Review.
-
A New Portrait of Constitutive Heterochromatin: Lessons from Drosophila melanogaster.Trends Genet. 2019 Sep;35(9):615-631. doi: 10.1016/j.tig.2019.06.002. Epub 2019 Jul 15. Trends Genet. 2019. PMID: 31320181 Review.
Cited by
-
The Drosophila gene encoding JIG protein (CG14850) is critical for CrebA nuclear trafficking during development.Nucleic Acids Res. 2023 Jun 23;51(11):5647-5660. doi: 10.1093/nar/gkad343. Nucleic Acids Res. 2023. PMID: 37144466 Free PMC article.
References
-
- Heitz E. Das geterochromatinder moose. J Jh Wiss. 1933;69:762–818.
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD et al. The Genome Sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
-
- Gatti M, Pimpinelli S. Cytological and genetical analysis of the Y chromosome of Drosophila melanogaster. . Chromosoma. 1983;88:593–617.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

