Regulation of DCC localization by HTZ-1/H2A.Z and DPY-30 does not correlate with H3K4 methylation levels
- PMID: 21998734
- PMCID: PMC3187824
- DOI: 10.1371/journal.pone.0025973
Regulation of DCC localization by HTZ-1/H2A.Z and DPY-30 does not correlate with H3K4 methylation levels
Abstract
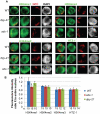
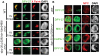
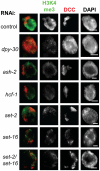
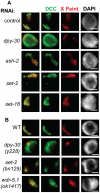
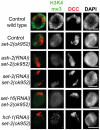
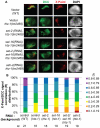
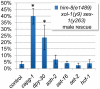
Dosage compensation is a specialized form of gene regulation that balances sex-chromosome linked gene expression between the sexes. In C. elegans, dosage compensation is achieved by the activity of the dosage compensation complex (DCC). The DCC binds along both X chromosomes in hermaphrodites to down-regulate gene expression by half, limiting X-linked gene products to levels produced in XO males. Sequence motifs enriched on the X chromosome play an important role in targeting the DCC to the X. However, these motifs are not strictly X-specific and therefore other factors, such as the chromatin environment of the X chromosome, are likely to aid in DCC targeting. Previously, we found that loss of HTZ-1 results in partial disruption of dosage compensation localization to the X chromosomes. We wanted to know whether other chromatin components coordinated with HTZ-1 to regulate DCC localization. One candidate is DPY-30, a protein known to play a role in DCC localization. DPY-30 homologs in yeast, flies, and mammals are highly conserved members of histone H3 lysine 4 (H3K4) methyltransferase Set1/MLL complexes. Therefore, we investigated the hypothesis that the dosage compensation function of DPY-30 involves H3K4 methylation. We found that in dpy-30 animals the DCC fails to stably bind chromatin. Interestingly, of all the C. elegans homologs of Set1/MLL complex subunits, only DPY-30 is required for stable DCC binding to chromatin. Additionally, loss of H3K4 methylation does not enhance DCC mislocalization in htz-1 animals. We conclude that DPY-30 and HTZ-1 have unique functions in DCC localization, both of which are largely independent of H3K4 methylation.
Conflict of interest statement
Figures
References
-
- Ercan S, Lieb JD. C. elegans dosage compensation: a window into mechanisms of domain-scale gene regulation. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2009;17:215–227. - PubMed
-
- Davis TL, Meyer BJ. SDC-3 coordinates the assembly of a dosage compensation complex on the nematode X chromosome. Development. 1997;124:1019–1031. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

