Transient, sparsely populated compact states of apo and calcium-loaded calmodulin probed by paramagnetic relaxation enhancement: interplay of conformational selection and induced fit
- PMID: 21999431
- PMCID: PMC3218299
- DOI: 10.1021/ja2082813
Transient, sparsely populated compact states of apo and calcium-loaded calmodulin probed by paramagnetic relaxation enhancement: interplay of conformational selection and induced fit
Abstract
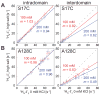
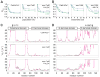
Calmodulin (CaM) is the universal calcium sensor in eukaryotes, regulating the function of numerous proteins. Crystallography and NMR show that free CaM-4Ca(2+) exists in an extended conformation with significant interdomain separation, but clamps down upon target peptides to form a highly compact structure. NMR has revealed substantial interdomain motions in CaM-4Ca(2+), enabled by a flexible linker. In one instance, CaM-4Ca(2+) has been crystallized in a compact configuration; however, no direct evidence for transient interdomain contacts has been observed in solution, and little is known about how large-scale interdomain motions contribute to biological function. Here, we use paramagnetic relaxation enhancement (PRE) to characterize transient compact states of free CaM that are too sparsely populated to observe by traditional NMR methods. We show that unbound CaM samples a range of compact structures, populated at 5-10%, and that Ca(2+) dramatically alters the distribution of these configurations in favor of states resembling the peptide-bound structure. In the absence of Ca(2+), the target peptide binds only to the C-terminal domain, and the distribution of compact states is similar with and without peptide. These data suggest an alternative pathway of CaM action in which CaM remains associated with its kinase targets even in the resting state. Only CaM-4Ca(2+), however, shows an innate propensity to form the physiologically active compact structures, suggesting that Ca(2+) activates CaM not only through local structural changes within each domain but also through more global remodeling of interdomain interactions. Thus, these findings illustrate the subtle interplay between conformational selection and induced fit.
Figures



References
-
- Yamniuk AP, Vogel HJ. Mol Biotechnol. 2004;27:33–57. - PubMed
-
- Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax A. Nature Struct Biol. 1995;2:768–776. - PubMed
-
- Chattopadhyaya R, Meador WE, Means AR, Quiocho FA. J Mol Biol. 1992;228:1177–1192. - PubMed
-
- Meador WE, Means AR, Quiocho FA. Science. 1992;257:1251–1255. - PubMed
-
- Zhang M, Tanaka T, Ikura M. Nature Struct Biol. 1995;2:758–767. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

