The Enzyme Function Initiative
- PMID: 21999478
- PMCID: PMC3238057
- DOI: 10.1021/bi201312u
The Enzyme Function Initiative
Abstract
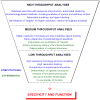
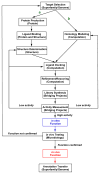
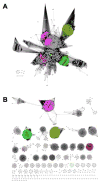
The Enzyme Function Initiative (EFI) was recently established to address the challenge of assigning reliable functions to enzymes discovered in bacterial genome projects; in this Current Topic, we review the structure and operations of the EFI. The EFI includes the Superfamily/Genome, Protein, Structure, Computation, and Data/Dissemination Cores that provide the infrastructure for reliably predicting the in vitro functions of unknown enzymes. The initial targets for functional assignment are selected from five functionally diverse superfamilies (amidohydrolase, enolase, glutathione transferase, haloalkanoic acid dehalogenase, and isoprenoid synthase), with five superfamily specific Bridging Projects experimentally testing the predicted in vitro enzymatic activities. The EFI also includes the Microbiology Core that evaluates the in vivo context of in vitro enzymatic functions and confirms the functional predictions of the EFI. The deliverables of the EFI to the scientific community include (1) development of a large-scale, multidisciplinary sequence/structure-based strategy for functional assignment of unknown enzymes discovered in genome projects (target selection, protein production, structure determination, computation, experimental enzymology, microbiology, and structure-based annotation), (2) dissemination of the strategy to the community via publications, collaborations, workshops, and symposia, (3) computational and bioinformatic tools for using the strategy, (4) provision of experimental protocols and/or reagents for enzyme production and characterization, and (5) dissemination of data via the EFI's Website, http://enzymefunction.org. The realization of multidisciplinary strategies for functional assignment will begin to define the full metabolic diversity that exists in nature and will impact basic biochemical and evolutionary understanding, as well as a wide range of applications of central importance to industrial, medicinal, and pharmaceutical efforts.
© 2011 American Chemical Society
Figures
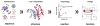

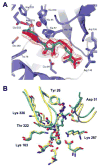
References
-
- Gerlt JA. A Protein Structure (or Function ?) Initiative. Structure. 2007;15:1353–1356. - PubMed
-
- Babbitt PC, Gerlt JA. Understanding enzyme superfamilies. Chemistry as the fundamental determinant in the evolution of new catalytic activities. J Biol Chem. 1997;272:30591–30594. - PubMed
-
- Gerlt JA, Babbitt PC. DIVERGENT EVOLUTION OF ENZYMATIC FUNCTION: Mechanistically Diverse Superfamilies and Functionally Distinct Suprafamilies. Annu Rev Biochem. 2001;70:209–246. - PubMed
-
- Gerlt JA, Babbitt PC, Rayment I. Divergent evolution in the enolase superfamily: the interplay of mechanism and specificity. Arch Biochem Biophys. 2005;433:59–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

