Recent advances in single-cell MALDI mass spectrometry imaging and potential clinical impact
- PMID: 21999830
- PMCID: PMC3268336
- DOI: 10.1586/epr.11.53
Recent advances in single-cell MALDI mass spectrometry imaging and potential clinical impact
Abstract
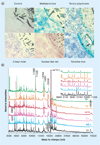
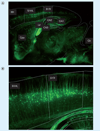
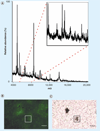
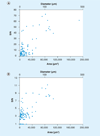
Single-cell analysis is gaining popularity in the field of mass spectrometry as a method for analyzing protein and peptide content in cells. The spatial resolution of MALDI mass spectrometry (MS) imaging is by a large extent limited by the laser focal diameter and the displacement of analytes during matrix deposition. Owing to recent advancements in both laser optics and matrix deposition methods, spatial resolution on the order of a single eukaryotic cell is now achievable by MALDI MS imaging. Provided adequate instrument sensitivity, a lateral resolution of approximately 10 µm is currently attainable with commercial instruments. As a result of these advances, MALDI MS imaging is poised to become a transformative clinical technology. In this article, the crucial steps needed to obtain single-cell resolution are discussed, as well as potential applications to disease research.
Figures




References
-
- McCombie G, Staab D, Stoeckli M, Knochenmuss R. Spatial and spectral correlations in MALDI mass spectrometry images by clustering and multivariate analysis. Anal. Chem. 2005;77(19):6118–6124. - PubMed
-
- Deininger SO, Ebert MP, Futterer A, Gerhard M, Rocken C. MALDI imaging combined with hierarchical clustering as a new tool for the interpretation of complex human cancers. J. Proteome Res. 2008;7(12):5230–5236. - PubMed
-
- Monroe EB, Jurchen JC, Lee J, Rubakhin SS, Sweedler JV. Vitamin E imaging and localization in the neuronal membrane. J. Am. Chem. Soc. 2005;127(35):12152–12153. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
