Emerging models of glutamate receptor ion channel structure and function
- PMID: 22000510
- PMCID: PMC3197216
- DOI: 10.1016/j.str.2011.08.009
Emerging models of glutamate receptor ion channel structure and function
Abstract
Excitatory synaptic transmission in the brain is mediated by ligand-gated ion channels (iGluRs) activated by glutamate. Distinct from other neurotransmitter receptors, the extracellular domains of iGluRs are loosely packed assemblies with two clearly distinct layers, each of which has both local and global 2-fold axes of symmetry. By contrast, the iGluR transmembrane segments have 4-fold symmetry and share a conserved pore loop architecture found in tetrameric voltage-gated ion channels. The striking layered architecture of iGluRs revealed by the 3.6 Å resolution structure of an AMPA receptor homotetramer likely arose from gene fusion events that occurred early in evolution. Although this modular design has greatly facilitated biophysical and structural studies on individual iGluR domains, and suggested conserved mechanisms for iGluR gating, recent work is beginning to reveal unanticipated diversity in the structure, allosteric regulation, and assembly of iGluR subtypes.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures



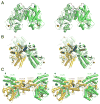

References
-
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: Crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. - PubMed
-
- Ayalon G, Segev E, Elgavish S, Stern-Bach Y. Two regions in the N-terminal domain of ionotropic glutamate receptor 3 form the subunit oligomerization interfaces that control subtype-specific receptor assembly. J Biol Chem. 2005;280:15053–15060. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

