Structural basis of Wnt signaling inhibition by Dickkopf binding to LRP5/6
- PMID: 22000856
- PMCID: PMC3215855
- DOI: 10.1016/j.devcel.2011.09.003
Structural basis of Wnt signaling inhibition by Dickkopf binding to LRP5/6
Abstract
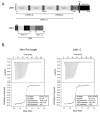
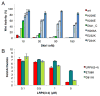
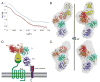
LDL receptor-related proteins 5 and 6 (LRP5/6) are coreceptors for Wnt growth factors, and also bind Dkk proteins, secreted inhibitors of Wnt signaling. The LRP5/6 ectodomain contains four β-propeller/EGF-like domain repeats. The first two repeats, LRP6(1-2), bind to several Wnt variants, whereas LRP6(3-4) binds other Wnts. We present the crystal structure of the Dkk1 C-terminal domain bound to LRP6(3-4), and show that the Dkk1 N-terminal domain binds to LRP6(1-2), demonstrating that a single Dkk1 molecule can bind to both portions of the LRP6 ectodomain and thereby inhibit different Wnts. Small-angle X-ray scattering analysis of LRP6(1-4) bound to a noninhibitory antibody fragment or to full-length Dkk1 shows that in both cases the ectodomain adopts a curved conformation that places the first three repeats at a similar height relative to the membrane. Thus, Wnts bound to either portion of the LRP6 ectodomain likely bear a similar spatial relationship to Frizzled coreceptors.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
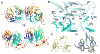


Comment in
-
Cell signaling. Crystallizing WNT signalling.Nat Rev Mol Cell Biol. 2012 Jan;13(1):4. doi: 10.1038/nrm3260. Nat Rev Mol Cell Biol. 2012. PMID: 22295277 No abstract available.
References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. - PubMed
-
- Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat Cell Biol. 2001;3:683–686. - PubMed
-
- Balemans W, Devogelaer JP, Cleiren E, Piters E, Caussin E, Van Hul W. Novel LRP5 missense mutation in a patient with a high bone mass phenotype results in decreased DKK1-mediated inhibition of Wnt signaling. J Bone Miner Res. 2007;22:708–716. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

