A novel hydrolase identified by genomic-proteomic analysis of phenylurea herbicide mineralization by Variovorax sp. strain SRS16
- PMID: 22003008
- PMCID: PMC3233098
- DOI: 10.1128/AEM.06162-11
A novel hydrolase identified by genomic-proteomic analysis of phenylurea herbicide mineralization by Variovorax sp. strain SRS16
Abstract
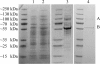
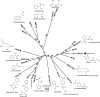
The soil bacterial isolate Variovorax sp. strain SRS16 mineralizes the phenylurea herbicide linuron. The proposed pathway initiates with hydrolysis of linuron to 3,4-dichloroaniline (DCA) and N,O-dimethylhydroxylamine, followed by conversion of DCA to Krebs cycle intermediates. Differential proteomic analysis showed a linuron-dependent upregulation of several enzymes that fit into this pathway, including an amidase (LibA), a multicomponent chloroaniline dioxygenase, and enzymes associated with a modified chlorocatechol ortho-cleavage pathway. Purified LibA is a monomeric linuron hydrolase of ∼55 kDa with a K(m) and a V(max) for linuron of 5.8 μM and 0.16 nmol min⁻¹, respectively. This novel member of the amidase signature family is unrelated to phenylurea-hydrolyzing enzymes from Gram-positive bacteria and lacks activity toward other tested phenylurea herbicides. Orthologues of libA are present in all other tested linuron-degrading Variovorax strains with the exception of Variovorax strains WDL1 and PBS-H4, suggesting divergent evolution of the linuron catabolic pathway in different Variovorax strains. The organization of the linuron degradation genes identified in the draft SRS16 genome sequence indicates that gene patchwork assembly is at the origin of the pathway. Transcription analysis suggests that a catabolic intermediate, rather than linuron itself, acts as effector in activation of the pathway. Our study provides the first report on the genetic organization of a bacterial pathway for complete mineralization of a phenylurea herbicide and the first report on a linuron hydrolase in Gram-negative bacteria.
Figures

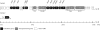


References
-
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 - PubMed
-
- Antoine R., et al. 2005. The periplasmic binding protein of a tripartite tricarboxylate transporter is involved in signal transduction. J. Mol. Biol. 351:799–809 - PubMed
-
- Breugelmans P., D'Huys P. J., De Mot R., Springael D. 2007. Characterization of novel linuron-mineralizing bacterial consortia enriched from long-term linuron-treated agricultural soils. FEMS Microbiol. Ecol. 62:374–385 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

