Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins
- PMID: 22003076
- PMCID: PMC3229142
- DOI: 10.1105/tpc.111.088377
Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins
Abstract
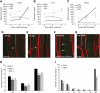
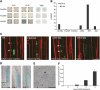
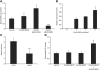
Multicellular organisms depend on cell production, cell fate specification, and correct patterning to shape their adult body. In plants, auxin plays a prominent role in the timely coordination of these different cellular processes. A well-studied example is lateral root initiation, in which auxin triggers founder cell specification and cell cycle activation of xylem pole-positioned pericycle cells. Here, we report that the E2Fa transcription factor of Arabidopsis thaliana is an essential component that regulates the asymmetric cell division marking lateral root initiation. Moreover, we demonstrate that E2Fa expression is regulated by the LATERAL ORGAN BOUNDARY DOMAIN18/LATERAL ORGAN BOUNDARY DOMAIN33 (LBD18/LBD33) dimer that is, in turn, regulated by the auxin signaling pathway. LBD18/LBD33 mediates lateral root organogenesis through E2Fa transcriptional activation, whereas E2Fa expression under control of the LBD18 promoter eliminates the need for LBD18. Besides lateral root initiation, vascular patterning is disrupted in E2Fa knockout plants, similarly as it is affected in auxin signaling and lbd mutants, indicating that the transcriptional induction of E2Fa through LBDs represents a general mechanism for auxin-dependent cell cycle activation. Our data illustrate how a conserved mechanism driving cell cycle entry has been adapted evolutionarily to connect auxin signaling with control of processes determining plant architecture.
Figures



Similar articles
-
PRH1 mediates ARF7-LBD dependent auxin signaling to regulate lateral root development in Arabidopsis thaliana.PLoS Genet. 2020 Feb 7;16(2):e1008044. doi: 10.1371/journal.pgen.1008044. eCollection 2020 Feb. PLoS Genet. 2020. PMID: 32032352 Free PMC article.
-
Effects of three auxin-inducible LBD members on lateral root formation in Arabidopsis thaliana.Planta. 2012 Oct;236(4):1227-37. doi: 10.1007/s00425-012-1673-3. Epub 2012 Jun 15. Planta. 2012. PMID: 22699776
-
The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins.Development. 2012 Mar;139(5):883-93. doi: 10.1242/dev.071928. Epub 2012 Jan 25. Development. 2012. PMID: 22278921
-
North, East, South, West: mapping vascular tissues onto the Arabidopsis root.Curr Opin Plant Biol. 2018 Feb;41:16-22. doi: 10.1016/j.pbi.2017.07.011. Epub 2017 Aug 30. Curr Opin Plant Biol. 2018. PMID: 28837854 Review.
-
Post-embryonic root organogenesis in cereals: branching out from model plants.Trends Plant Sci. 2013 Aug;18(8):459-67. doi: 10.1016/j.tplants.2013.04.010. Epub 2013 May 31. Trends Plant Sci. 2013. PMID: 23727199 Review.
Cited by
-
Lateral Organ Boundaries Domain16 and 18 Act Downstream of the AUXIN1 and LIKE-AUXIN3 Auxin Influx Carriers to Control Lateral Root Development in Arabidopsis.Plant Physiol. 2015 Aug;168(4):1792-806. doi: 10.1104/pp.15.00578. Epub 2015 Jun 9. Plant Physiol. 2015. PMID: 26059335 Free PMC article.
-
Arabidopsis ULTRAVIOLET-B-INSENSITIVE4 maintains cell division activity by temporal inhibition of the anaphase-promoting complex/cyclosome.Plant Cell. 2011 Dec;23(12):4394-410. doi: 10.1105/tpc.111.091793. Epub 2011 Dec 13. Plant Cell. 2011. PMID: 22167059 Free PMC article.
-
Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes.EMBO J. 2012 Mar 21;31(6):1480-93. doi: 10.1038/emboj.2012.13. Epub 2012 Feb 3. EMBO J. 2012. PMID: 22307083 Free PMC article.
-
CYTOKININ RESPONSE FACTOR2 (CRF2) and CRF3 Regulate Lateral Root Development in Response to Cold Stress in Arabidopsis.Plant Cell. 2016 Aug;28(8):1828-43. doi: 10.1105/tpc.15.00909. Epub 2016 Jul 18. Plant Cell. 2016. PMID: 27432872 Free PMC article.
-
PRH1 mediates ARF7-LBD dependent auxin signaling to regulate lateral root development in Arabidopsis thaliana.PLoS Genet. 2020 Feb 7;16(2):e1008044. doi: 10.1371/journal.pgen.1008044. eCollection 2020 Feb. PLoS Genet. 2020. PMID: 32032352 Free PMC article.
References
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57: 289–300
-
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 - PubMed
-
- Berckmans B., De Veylder L. (2009). Transcriptional control of the cell cycle. Curr. Opin. Plant Biol. 12: 599–605 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

