The regulatory activities of plant microRNAs: a more dynamic perspective
- PMID: 22003084
- PMCID: PMC3327222
- DOI: 10.1104/pp.111.187088
The regulatory activities of plant microRNAs: a more dynamic perspective
Figures
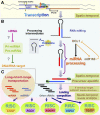
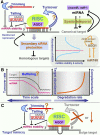
References
-
- Allen E, Howell MD. (2010) miRNAs in the biogenesis of trans-acting siRNAs in higher plants. Semin Cell Dev Biol 21: 798–804 - PubMed
-
- Allen E, Xie Z, Gustafson AM, Carrington JC. (2005) MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 - PubMed
-
- Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC. (2004) Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet 36: 1282–1290 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

