Adenovirus gene transfer to amelogenesis imperfecta ameloblast-like cells
- PMID: 22003382
- PMCID: PMC3189176
- DOI: 10.1371/journal.pone.0024281
Adenovirus gene transfer to amelogenesis imperfecta ameloblast-like cells
Abstract
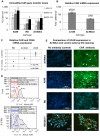
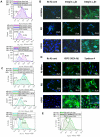
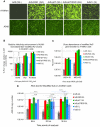
To explore gene therapy strategies for amelogenesis imperfecta (AI), a human ameloblast-like cell population was established from third molars of an AI-affected patient. These cells were characterized by expression of cytokeratin 14, major enamel proteins and alkaline phosphatase staining. Suboptimal transduction of the ameloblast-like cells by an adenovirus type 5 (Ad5) vector was consistent with lower levels of the coxsackie-and-adenovirus receptor (CAR) on those cells relative to CAR-positive A549 cells. To overcome CAR -deficiency, we evaluated capsid-modified Ad5 vectors with various genetic capsid modifications including "pK7" and/or "RGD" motif-containing short peptides incorporated in the capsid protein fiber as well as fiber chimera with the Ad serotype 3 (Ad3) fiber "knob" domain. All fiber modifications provided an augmented transduction of AI-ameloblasts, revealed following vector dose normalization in A549 cells with a superior effect (up to 404-fold) of pK7/RGD double modification. This robust infectivity enhancement occurred through vector binding to both α(v)β3/α(v)β5 integrins and heparan sulfate proteoglycans (HSPGs) highly expressed by AI-ameloblasts as revealed by gene transfer blocking experiments. This work thus not only pioneers establishment of human AI ameloblast-like cell population as a model for in vitro studies but also reveals an optimal infectivity-enhancement strategy for a potential Ad5 vector-mediated gene therapy for AI.
Conflict of interest statement
Figures


References
-
- Simmer JP, Fincham AG. Molecular mechanisms of dental enamel formation. Crit Rev Oral Biol Med. 1995;6:84–108. - PubMed
-
- Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU. Properties of dissociatively extracted fetal tooth matrix proteins. I. Principal molecular species in developing bovine enamel. J Biol Chem. 1980;255(20):9760–8. - PubMed
-
- Zalzal SF, Smith CE, Nanci A. Ameloblastin and amelogenin share a common secretory pathway and are co-secreted during enamel formation. Matrix Biol. 2008;27(4):352–9. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

